Primary sequencing output of an Illumina sequencer are per-cycle base call (bcl) files, which first need to be converted to fastq format, so that the data can be pushed to downstream applications. The conversion tool implemented in Partek Flow is Illumina's bcl2fastq Conversion Software. In addition to the file conversion, bcl2fastq tool also demultiplexes the bcl files in the same step and, hence, outputs demultiplexed fastq files as the primary result. The bcl2fastq application also supports two optional tasks: adapter trimming and removal of unique molecular identifier (UMI) sequences.
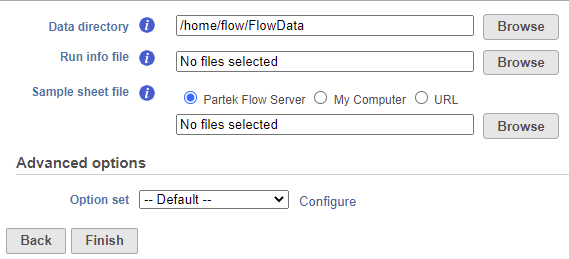
To base a new project on bcl files, first select Import data option on the Data tab and then push the Import bcl files button. The resulting window shows the configuration dialog (Figure 1).
The RunInfo.xml file is generated by the primary analysis software and contains information on the run, flow cell, the instrument, the time stamp, and the read structure (number of reads, number of cycles per read, whether a read is an index read). The SampleSheet.csv file provides the information on the relationship between the samples and indices specified during library creation. It has four sections, but bcl2fastq tool uses only two of those (Settings and Data). Finally, the bcl files hold the base calls and are contained within the BaseCalls directory (note that the Select base calls directory option needs to point to the directory, not to an individual bcl file). For more information on the files, consult Illumina documentation.
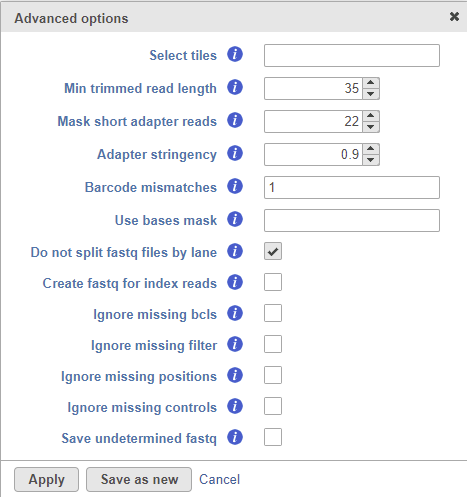
Selecting the Configure option under Advanced options section enables a granular control of the import (Figure 2). The Select tiles option (--tiles) enables the user to process only a subset of tiles available in the flow cell. The input for this option is a comma-separated list of regular expressions. Min trimmed read length (--minimum-trimmed-read-length) specifies the minimum read length after adapter removal. Mask short adapter reads (--mask-short-adapter-reads) applies when a read is trimmed below the length specified by Min trimmed read length. If the number of bases after adapter removal is less than Min trimmed read length, it forces the read length to be equal to Min trimmed read length by replacing the adapter bases that fall below the specified length by Ns. If the number of remaining bases falls below Mask short adapter sequences, then replace all the bases in a read with Ns. Adapter stringency (--adapter-stringency) specifies the minimum match rate that triggers the masking or trimming of adapters. The rate is calculated as MatchCount / (MatchCount + MismatchCount). Only the reads exceeding the specified rate of sequence identity with adapters are trimmed. Barcode mismatches (--barcode-mismatches) controls the number of allowed mismatches per index sequence. Do not split files by lane (--no-lane-splitting) prevents splitting of fastq files by lane, i.e. the converter will merge multiple lanes and generate one fastq file per sample.