...
You can invoke the creation of the plot on the annotated peak data node. After the peaks are annotated with transcripts, coverage of the reads across the genes overlapped with the regions will be calculated, this information is presented by profile plot (Figure 1) and heatmap (Figure 4).
Select the Annotated regions results node then under Exploratory analysis in the task menu, select TSS plot.
| Table of Contents |
|---|
| maxLevel | 2 |
|---|
| minLevel | 2 |
|---|
| exclude | Additional Assistance |
|---|
|
| Numbered figure captions |
|---|
| SubtitleText | TSS profile plot |
|---|
| AnchorName | tss_profile |
|---|
|
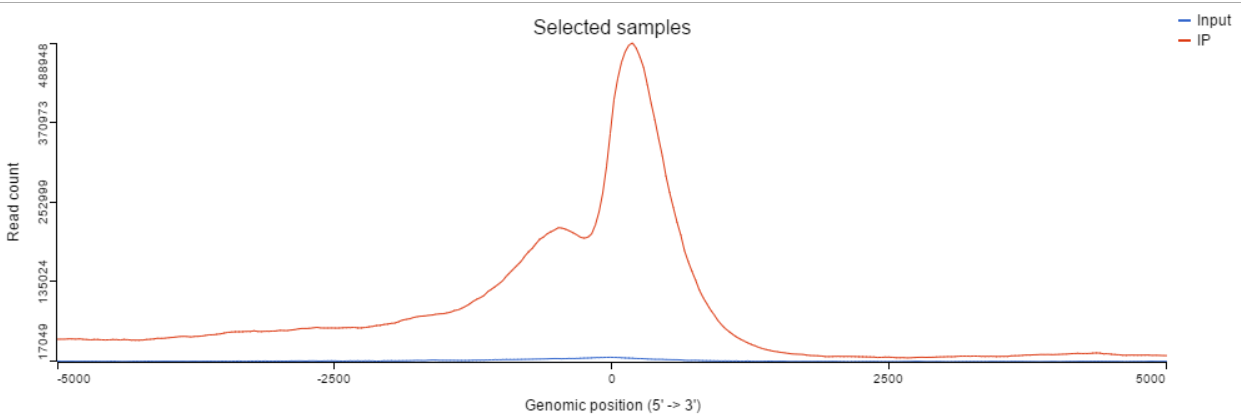 Image Modified Image Modified
|
TSS profile plot
In the profile plot, the Y-axis is the total read counts, X-axis is the window defined in the annotation task, mid point is the transcription start site (TSS) of all the genes overlapping with the peak regions, the start and end point of X-axis is defined by the promoter upstream/downstream limit specified in the annotate peaks task (Figure 2). Each line represents a selected sample total read counts at each position, all the selected samples are shown in the plot to compare.
| Numbered figure captions |
|---|
| SubtitleText | Annotate peaks task dialog |
|---|
| AnchorName | annotation |
|---|
|
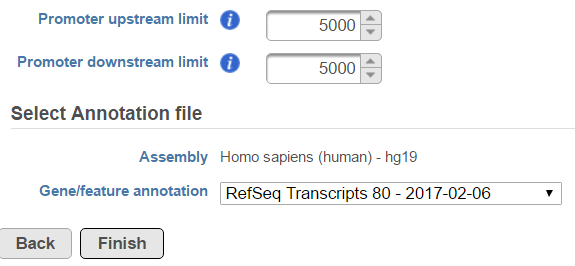 Image Modified Image Modified
|
By default, all the samples are selected to display, to remove a sample track, click on the  button next to the sample name on the sample control panel (Figure 3); to add a sample in the plot, select the sample name from the Add sample drop-down list, a new line will be added on the plot.
button next to the sample name on the sample control panel (Figure 3); to add a sample in the plot, select the sample name from the Add sample drop-down list, a new line will be added on the plot.
| Numbered figure captions |
|---|
| SubtitleText | Sample selection control panel |
|---|
| AnchorName | select_sample |
|---|
|
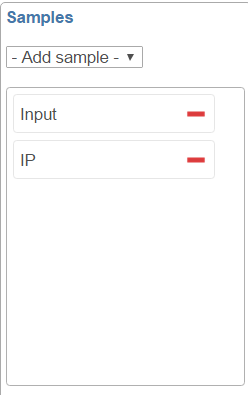 Image Modified Image Modified
|
TSS heatmap
The heatmap is another way to view the overlap between the gene body and peaks detected (Figure 4). The X-axis represents the same information as the profile plot. While the maximum value of the Y-axis varies among different samples. In the heatmap, each row represents a gene, while the color encodes the total read count at each position and the genes are sorted based on total read counts in descending order.
| Numbered figure captions |
|---|
| SubtitleText | TSS heatmap |
|---|
| AnchorName | tss_heatmap |
|---|
|
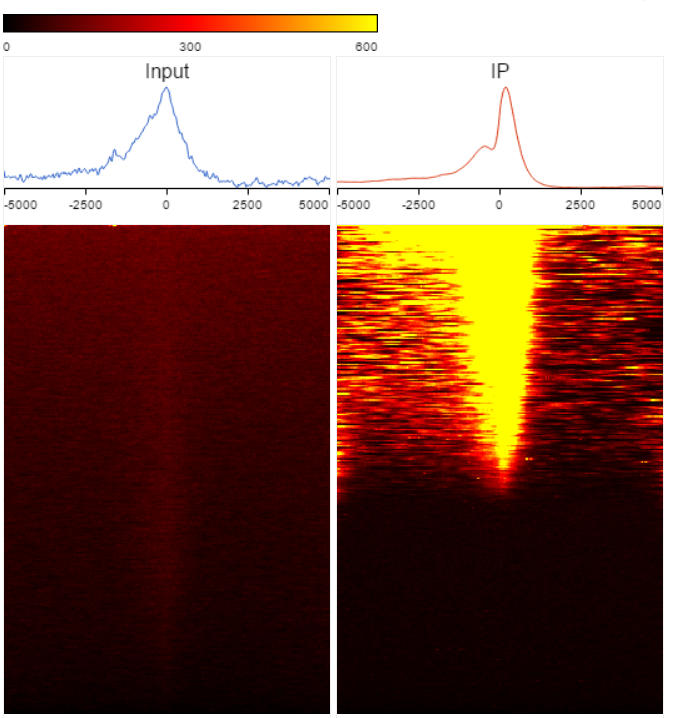 Image Modified Image Modified
|
The color and color scale on the heatmap can be customized using the control panel on the left (Figure 5). You can change the number of heatmaps per row, with a higher number decreasing the heatmap size. For the heat map colors, changing the Low and High values will change the color scale of the heatmap. You can also change the color of the profile line by selecting Customize sample colors.
| Numbered figure captions |
|---|
| SubtitleText | TSS heatmap control panel |
|---|
| AnchorName | heatmap_control |
|---|
|
 Image Modified Image Modified
|
...
...
|
...





