Page History
...
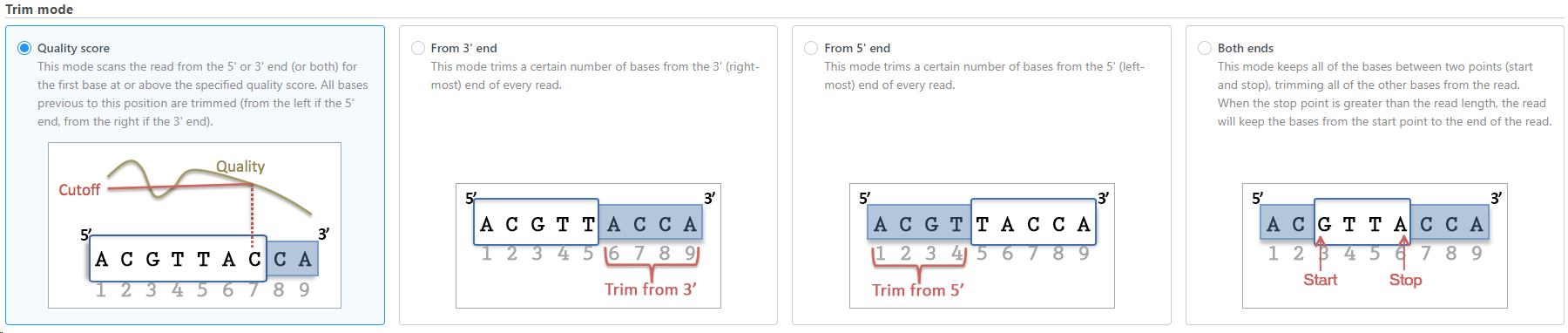
The task allows user to trim reads in different ways (Figure 1), including:
- Trim bases based on quality score
- Trim bases from 3'-end
- Trim bases from 5'-end
- Trim bases from both ends
| Numbered figure captions | ||
|---|---|---|
|
...
| |||
Trim bases from 5'-or 3'-end (Figures 12-23) allows a fixed number of bases to be trimmed away from the 5'- or 3'-end of the reads. These two functions are useful for when your read length is constant. This is not recommended if the read length is not constant, since good quality bases from shorter reads are likely trimmed away by these functions.
...
Trim bases from both ends (Figure 34) allows user to keep only bases from a fixed start and end position of the reads. This is particularly useful if poor quality bases are observed on both ends of the read. So instead of performing trim bases successively from the 5'- and 3'-end, the trim bases will only be performed once by trimming from both ends.
...
Trim bases based on quality score (Figure 45) is probably the most useful function to trim poor quality bases from the 5'- or 3'-ends of reads. This function allows dynamic trimming of bases depending on quality score. The trimming can be done from either 5'-end, 3'-end or both ends of the reads. The function evaluates each base from the end of the read and trims it away until the last base has a quality score greater than the specified threshold. For an extensive evaluation of read trimming effects on Illumina NGS data analysis, see Del Fabbro et. al. [1].
...
The Quality encoding option refers to the Phred quality score encoded within the FASTQ input file. The list of available options are: Phred+33, Phred+64, Solexa+64 and Integers. Selecting Auto-detect will determine whether the quality encoding is Phred+33 or Phred+64. For Solexa data, you will need to select Solexa+64. For most of datasets, auto-detect option works very well with a few exception cases where the base quality score falls into the grey zone (ambiguous zone) of Phred+33 and Phred+64 score. However, if the quality-encoding scheme is known, we recommend to selecting the encoding format directly from the quality encoding list.
Figure 5 6 shows the options available for all the different selection of Trim bases function. Note the default Min read length is 25bp. For micro RNA sequencing data, this default Min read length needs to be set to a smaller value (we recommend 15) to account for mature microRNAs.
...