Page History
Missing library files can be added when setting up tasks within a project, without having to navigate to the library file management page. The user interface will vary depending on the task and which library files already exist on your system. Below are two examples scenarios.
Adding a missing whole genome aligner index (Bowtie 2 in this example)
- Under the Analyses tab of a project, select an Unaligned reads data node
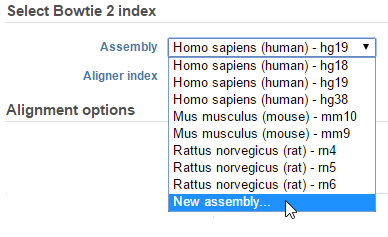
- From the context sensitive menu on the right, choose Aligners followed by Bowtie 2. On the alignment task setup page, Partek Flow will display all assemblies that have a Bowtie 2 index (whole genome and transcriptome) in the Assembly drop-down list. If the assembly you want is missing, choose New assembly… from the drop-down list (Figure 1)
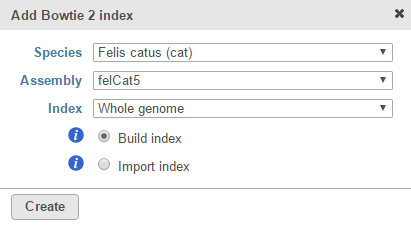
Figure 1. The drop-down list will show all assemblies that have a Bowtie 2 index associated with them. Choose New assembly... to add a Bowtie 2 index to another assembly - Choose the species and assembly in the Add Bowtie 2 index dialog. If the species and assembly you want do not appear in the drop-down lists, choose Other and manually type the names (Figure 2)
- Choose Whole genome from the Index drop-down list (Figure 2)
- Select the Build index radio button (Figure 2)
Click Create (Figure 2)
...
Once the new Bowtie 2 index has been specified, you are able to queue the alignment task and it will execute once the Bowtie 2 index has been built.
Add a missing gene set file for
...
enrichment analysis
- Under the Analyses tab of a project, select a Feature list data node
- Choose Biological interpretation from the menu on the right, followed by Enrichment analysis
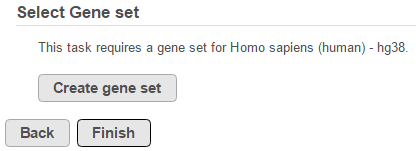
If there are no gene set files associated with the relevant assembly, click Create gene set (Figure 3)
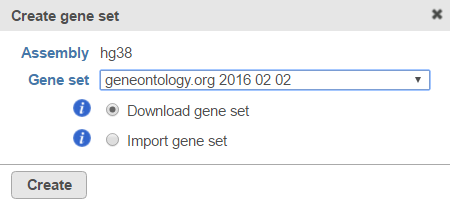
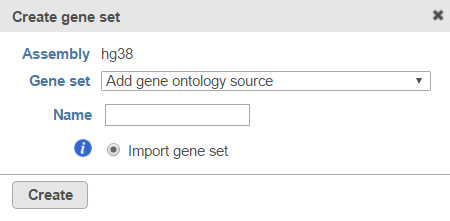
Figure 3. If no gene sets are associated with the assembly, click the button to add one.If you are working with an assembly/species supported by Partek (e.g. human), choose a gene set from the Create gene set drop-down list (Figure 4), select the Download gene set radio button and select Create. Alternatively, choose Add gene ontology source from the Create gene set drop-down list, manually type the custom gene set name and click Create to import your own gene set from the Partek Flow server, My computer or URL (Figure 5). Characters such as $ * | \ : " < > ? / % cannot be used in custom names. If you are working with a custom species/assembly (e.g. for a non-model organism), only the Add gene ontology source option is available.
Figure 4. Download a gene set using the Create gene set dialog from within a projectFigure 5. Import a gene set using the Create gene set dialog from within a project
...