Page History
...
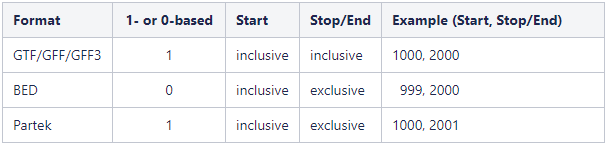
Genome coordinates for annotation models stored in Partek Flow are 1-based, start-inclusive, and stop-exclusive. This means that the first base position starts from one, the start coordinate for a feature is included in the feature and the stop/end coordinate is not included in the feature. These are the genome coordinates that are printed in various task reports and output files when an annotation model is involved in the task. When custom annotation files are added to Partek Flow, the genome coordinates are converted into this format. The coordinates are converted back if necessary for a specific task. The table below Figure 5 shows how the genome coordinates vary between different annotation formats. The
Figure 5. Comparison of genome coordinate formats. The example column shows the coordinates for a feature that starts at position 1000 (inclusive) and stops at position 2000 (inclusive).
...
| Additional assistance |
|---|
| Page Turner | ||
|---|---|---|
|
...