Page History
...
Selecting MACS2 from the context sensitive menu will bring up the MACS2 task dialog, interface will be different depends the input aligned data node.
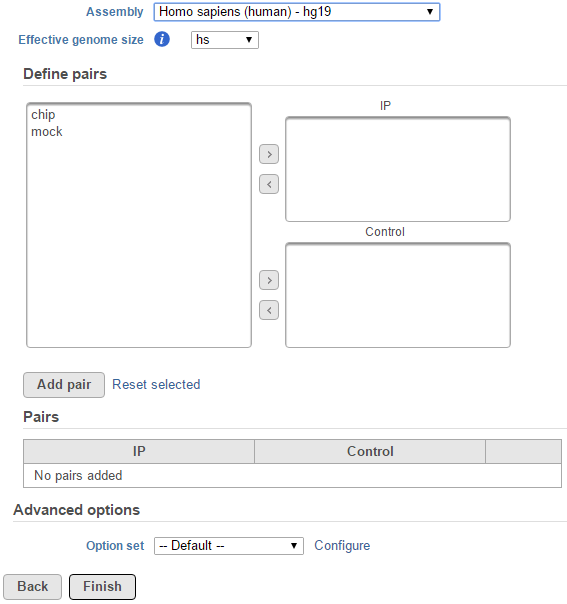
whether there are sample attributes available in the data tab. If there are only two sample for instance, ChIP and mock as sample name, there is no attribute assigned to the two samples, the dialog will look like (Figure 1)
If the selected aligned data node was imported, reference assembly the data aligned to needs to be specified, choose the assembly the sequence aligned to from the drop-down list (Figure 1); if the alignment was performed in Partek Flow, this option will not appear.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
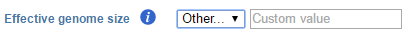
The effective genome size is the genome size can be sequenced. Because of the repetitive features on the chromosomes, the actual mappable genome size will be smaller than the original size, typically about 70%-90% of the genome size. There are presets of 4 species based on MACS2 recommendation1 for this parameter:
hs – human, size is 2.7e9
mm – mouse, size is 1.87e9
ce – C. elegans, size is 9e7
dm – fruit fly, size is 1.2e8
When Other is selected, a specific value of the effective genome size needs to be specified in bps as unit (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
When sample attribute is not specified, for instance there are only two sample -- ChIP and mock as sample name, the peak detection pairs needs to be manually defined (Figure 1).
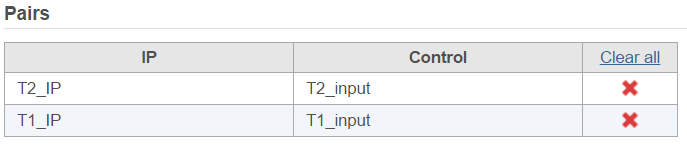
In the Define pairs section, the left panel list all the sample names, add one pair at a time, select ChIP sample to put in IP panel on the top-right, choose control sample to put in the Control panel on the bottom-right. If there is no control sample in the experiment, the Control panel can be blank. If more than one ChIP or Control samples added, the samples will be combined (or pooled) in the analysis.
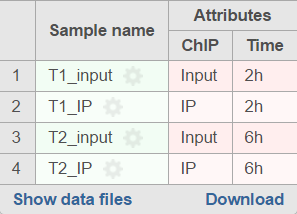
If the sample attributes are defined, you will have an additional option to add pairs based on the attribute. For instance Figure 2 3 is show an example data with 4 samples, 2 time point, there in one ChIP sample and one input sample in each time point.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
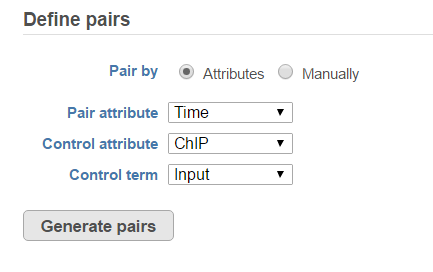
When select MACS2 task, the default option is to use sample attribute to add multiple pairs at one button click (Figure 34)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
There are IP-Input pair in each time point, so the pair attribute is Time; Control attribute is the attribute contains IP and input group, which is ChIP, the control term is labeled as Input in the example, when click Generate pairs, the two pairs will be automatically added to the Pairs table at once (Figure 45).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Peaks report
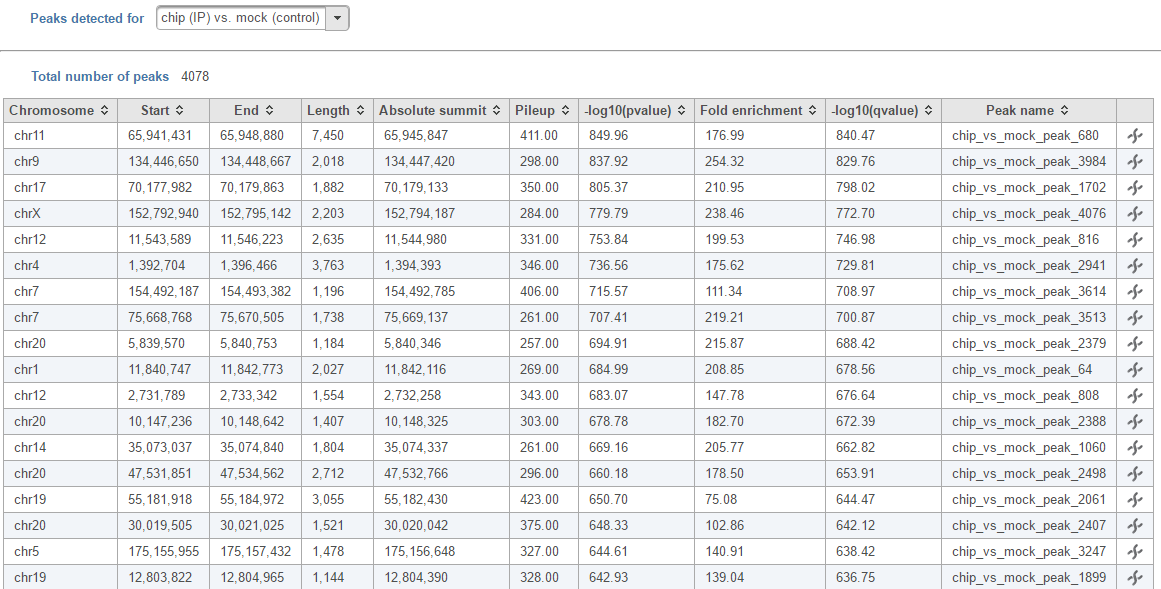
The peaks report is generated on each pair specified in the Pairs table separately (Figure 4In the task report, each pair will generate a list of peaks displayed in a table (Figure 6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
Absolute summit: base pair location of peak summit
Pileup: pileup height at peak summit
-log10(pvalue): negative log10 pvalue for thhe peak summit
Fold enricment: fold enrichhment for thhe peak summit against random Poisson distribution with local lambda
-log10(qvalue): negative log10 qvalue at peak summit
Click on the browse to peak button () to invoke chromosome view and zoom into that location.
References
- Zhang Y, Liu T, et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137.
...