Page History
...
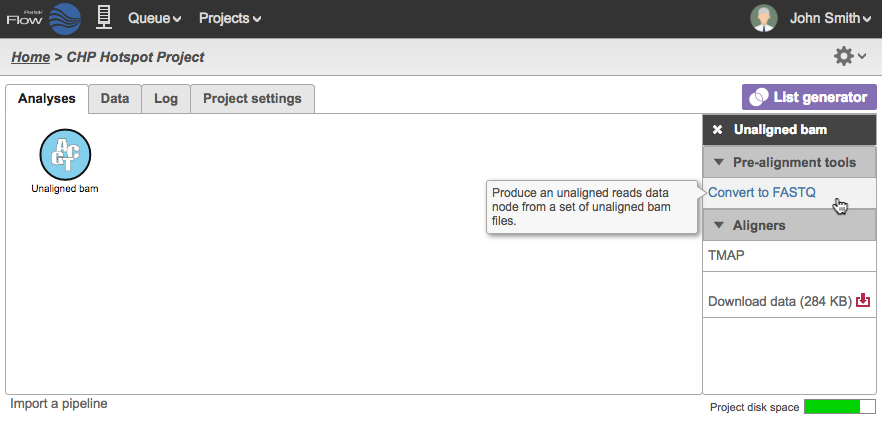
The plugin transfers the Unaligned BAM data from the Torrent Browser. The UBAM file format retains all the information of the Ion Torrent Sequencer. In the Partek Flow Project, the Analyses tab would show a circular data node named Unaligned bam. Click on the data node and the context-sensitive task menu will appear on the right (Figure 23).
Unaligned BAM files are only compatible with the TMAP aligner, which can be selected in the Aligners section of the Task Menu. If you wish to use other aligners, you can convert the unaligned BAM files to FASTQ using the Convert to FASTQ task under Pre-alignment tools.
Some information specific to Ion Torrent Data (such as Flow Order) are not retained in the FASTQ format. However, those are only relevant to Ion Torrent developed tools (such as the Torrent Variant Caller) and are not relevant to any other analysis tools.
Figure 23: Analyses tab of Partek Flow showing the task menu available for the selected Unaligned BAM file data node
Once converted, the reads can then be aligned using a variety of aligners compatible with fastq input (Figure 24) . You can also perform other tasks such as Pre-alignment QAQC or run an existing pipeline.
Another option is to include the Convert to FASTQ task in your pipeline and you can invoke the pipeline directly from an Unaligned bam data node.
Figure 24: Unaligned reads in the FASTQ format are compatible with more tasks
...