Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Import FASTQs for Cell Ranger -ATAC in Flow
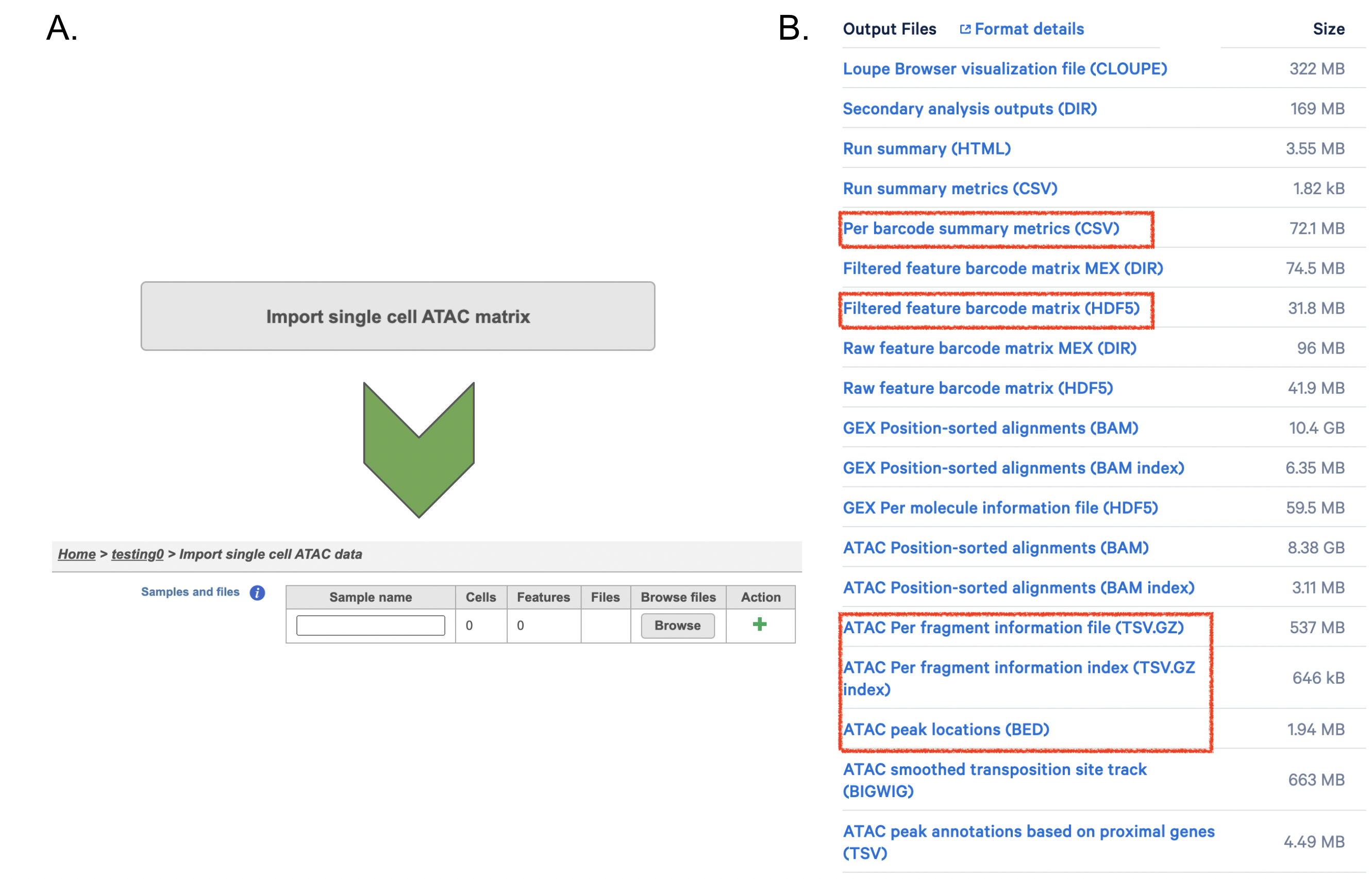
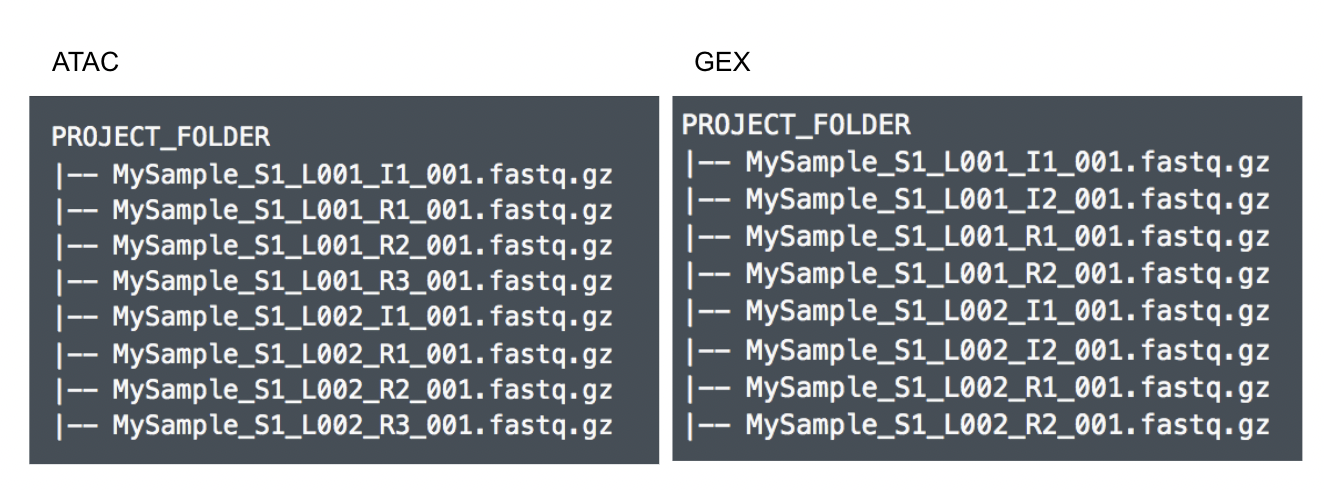
Although the index files (I1 or I2) are optional, we encourage users to include all the FASTQs in the table (Figure 9) while importing data for Cell Ranger - ATAC.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
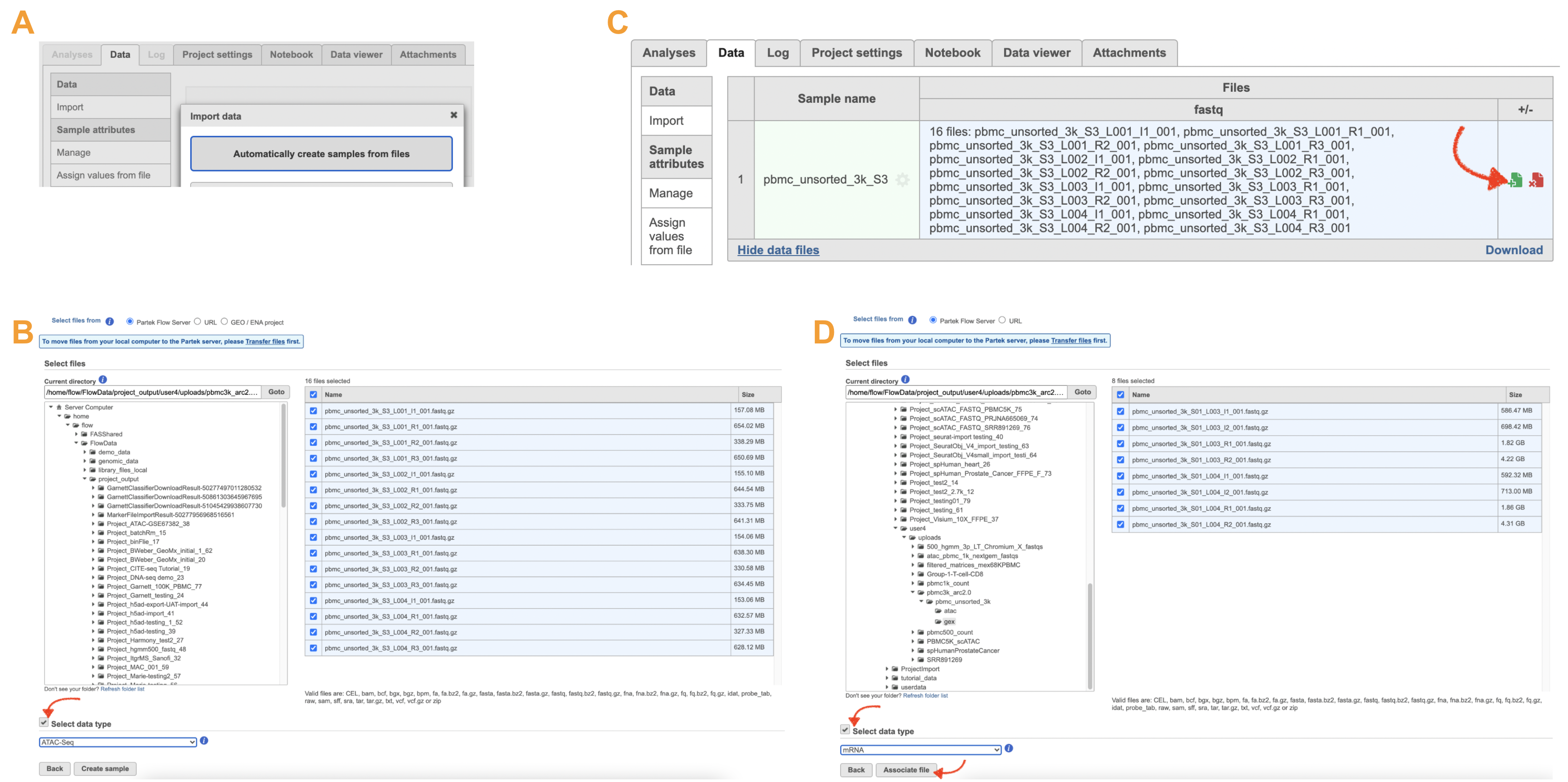
Import ATAC FASTQ files are as straightforward as the (sc)RNA-seq data. However, we have to associate two different types of data types together for 10x multiome ATAC + Gene Expression data. It includes two steps: 1). Import ATAC FASTQ files in the new page after clicking on the "Automatically create samples from files" in Data tab (Figure 10A) and select data type from the dropdown list (Figure 10B) to be ATAC-Seq. 2). Move back to the Data tab and display all the files by clicking on the Show data files button at the bottom left of Sample name table. Then go clicking the green + button (Figure 10C)to add RNA FASTQs to the same sample. Similarly, we will select data type from the dropdown (mRNA) before we finish the import process by clicking on the Associate file button (Figure 10D).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
References
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
https://support.10xgenomics.com/single-cell-atac/software/pipelines/latest/release-notes
- https://support.10xgenomics.com/single-cell-multiome-atac-gex/software/pipelines/latest/what-is-cell-ranger-arc
- https://support.10xgenomics.com/single-cell-multiome-atac-gex/software/pipelines/latest/output/summary
...