...
| Numbered figure captions |
|---|
| SubtitleText | Specifying attribute levels for sub-group comparisons (contrast): Select A for Cell type on the top, B for Cell type on the bottom, and click Add comparison to compare A vs B |
|---|
| AnchorName | Subgroup attribute selection |
|---|
|
 Image Modified Image Modified
|
To compare Time point 5 vs. 0, select 5 for Time on the top, 0 for Time on the bottom, and click Add comparison (Figure 3).
...
| Numbered figure captions |
|---|
| SubtitleText | Specifying attribute levels for sub-group comparisons (contrast): Select 5 for Time on the top, 0 for Time on the bottom, click Add comparison to compare 5 vs 0 |
|---|
| AnchorName | Subgroup Attribute comparison |
|---|
|
 Image Modified Image Modified
|
To compare cell types at a certain time point, e.g. time point 5, select A and 5 on the top, and B and 5 on the bottom. Thereafter click Add comparison (Figure 4).
...
| Numbered figure captions |
|---|
| SubtitleText | Specifying attribute levels for subgroup comparisons (contrast): Select A and 5 on the top, B and 5 on the bottom, click Add comparison to compare A*5 vs B*5 |
|---|
| AnchorName | Subgroup attribute comparrison contrast |
|---|
|
 Image Modified Image Modified
|
Multiple comparisons can be computed in one GSA run; Figure 5 shows the above three comparisons are added in the computation.
...
| Numbered figure captions |
|---|
| SubtitleText | Configuring advanced GSA options |
|---|
| AnchorName | GSA advanced options |
|---|
|
 Image Modified Image Modified
|
Low-expression feature
...
| Numbered figure captions |
|---|
| SubtitleText | Volcano plot in comparison A vs B. X-axis represents fold change (linear scale), Y-axis represents negative logged p-value (unadjusted), each dot is a feature. The horizontal line represents p-value of 0.05, two vertical lines represent fold change of -2 and 2. Lower left corner displays number of features passing the fold-change and p-value criteria |
|---|
| AnchorName | Volcano plot |
|---|
|
 Image Modified Image Modified
|
Feature list filter panel is on the left of the table (Figure 12). Click on the black triangle (  ) to collapse and expand the panel.
) to collapse and expand the panel.
Select the check box of the field and specify the cutoff, and press Enter to apply. After the filter has been applied, the total number of included features will be updated on the top of the panel (Result).
| Numbered figure captions |
|---|
| SubtitleText | Feature list filter panel |
|---|
| AnchorName | Feature list filter panel |
|---|
|
 Image Modified Image Modified
|
The filtered result can be saved into a filtered data node by selecting the Generate list button at the lower-left corner of the table (  ). Selecting the Download button at the lower-right corner of the table downloads the table as a text file to the local computer.
). Selecting the Download button at the lower-right corner of the table downloads the table as a text file to the local computer.
...
| Numbered figure captions |
|---|
| SubtitleText | ANOVA comparisons setup dialog: The example in the figure shows a single factor (Cell type) with two levels (A and B). A contrast A vs. B has been set |
|---|
| AnchorName | Comparisons setup dialog |
|---|
|
 Image Modified Image Modified
|
Start by choosing a factor or interaction from the Factor drop-down list. The subgroups of the factor or interaction will be displayed in the left panel; click to select a subgroup name and move it to one of the panels on the right. The fold change calculation on the comparison will use the group in the top panel as numerator, and the group in the bottom panel as the denominator. Click on Add comparison button to add one comparison to the comparisons table. Note that multiple comparisons can be added to the specified model.
...
| Numbered figure captions |
|---|
| SubtitleText | Configuring advanced options when running ANOVA |
|---|
| AnchorName | Advanced ANOVA options |
|---|
|
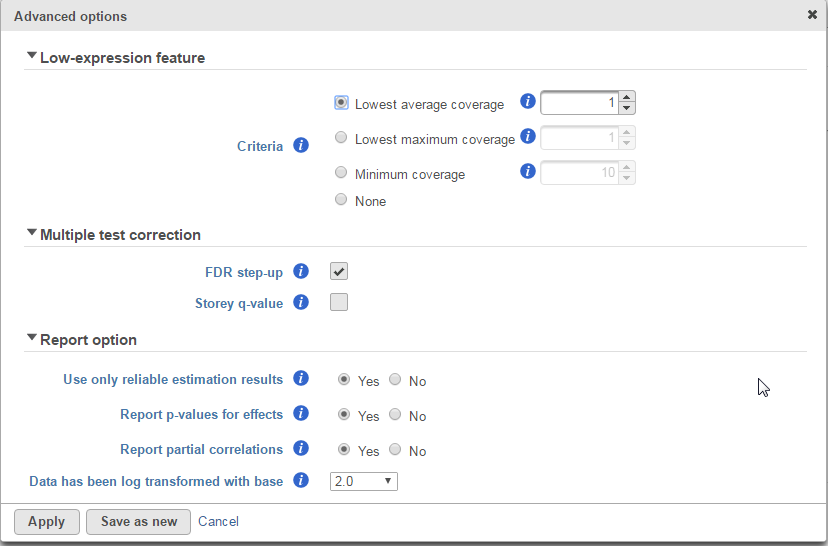 Image Modified Image Modified
|
Low-expression feature and Multiple test correction sections are the same as the matching GSA advanced option, see above GSA advanced options.
...
| Numbered figure captions |
|---|
| SubtitleText | Advanced option of cuffdiff |
|---|
| AnchorName | Advanced options of cuffdiff |
|---|
|
 Image Modified Image Modified
|
There are three library normalization methods:
...
- Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing, JRSS, B, 57, 289-300.
- Storey JD. (2003) The positive false discovery rate: A Bayesian interpretation and the q-value. Annals of Statistics, 31: 2013-2035.
- Auer, 2011, A two-stage Poisson model for testing RNA-Seq
- Burnham, Anderson, 2010, Model selection and multimodel inference
- Law C, Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biology, 2014 15:R29.
- http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html#cuffdiff-output-files
- Anders S, Huber W: Differential expression analysis for sequence count data. Genome Biology, 2010