Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
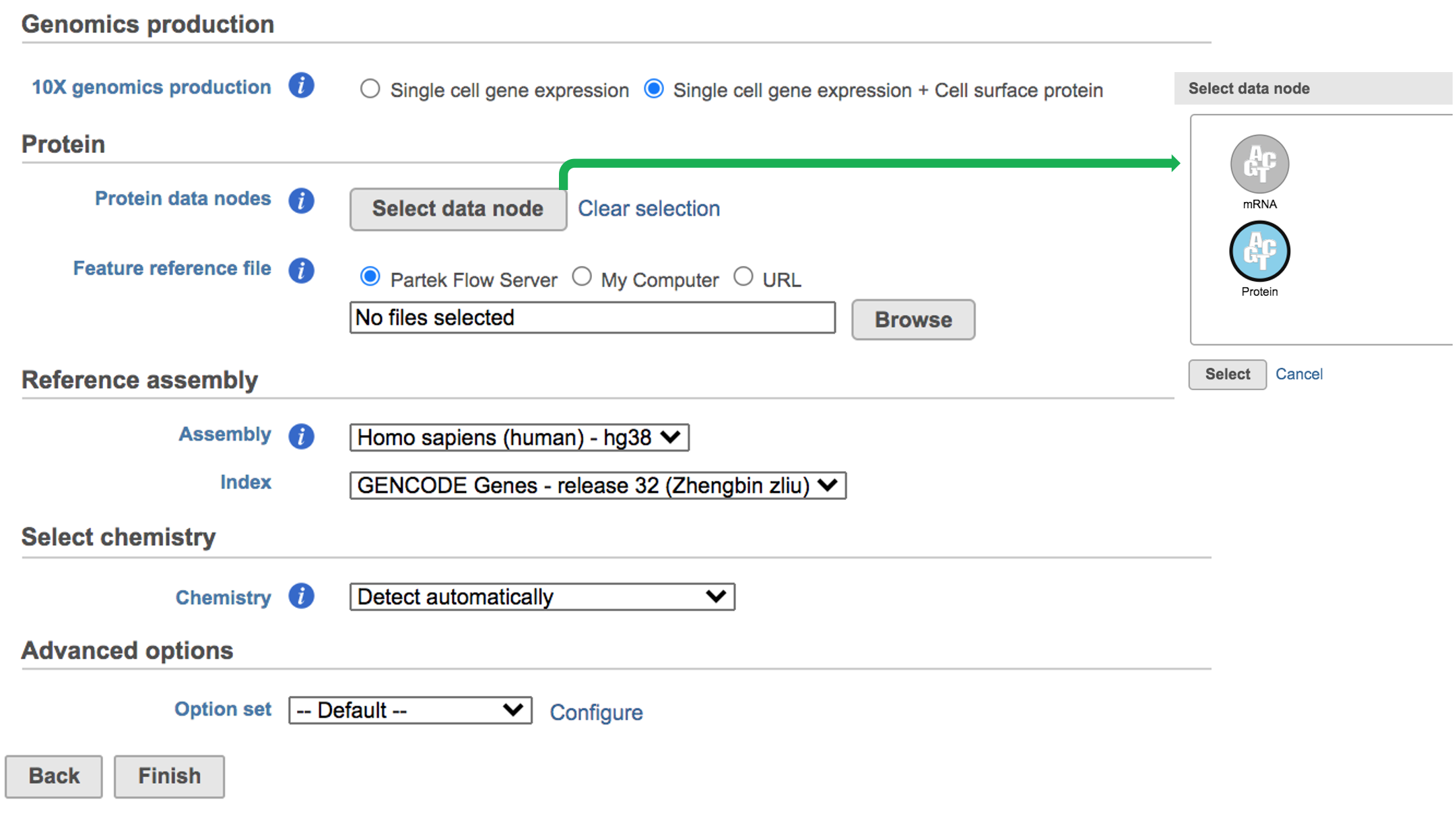
A new data node named ‘Single Single cell counts’ will will be displayed in Flow if the task has been finished successfully (Figure 6). This data node contains a filtered feature barcode count matrix for gene expression data, but a unified feature-barcode matrix that contains gene expression counts alongside Feature Barcode counts for each cell barcode for Feature Barcode data. To open the task report when the task is finished, double click the output data node, or select the ‘Task report’ in in the Task results section after single clicking the data node. Users then will find the task report (Figure 7) is the same to the ‘Summary HTML’ from Cell Ranger output.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
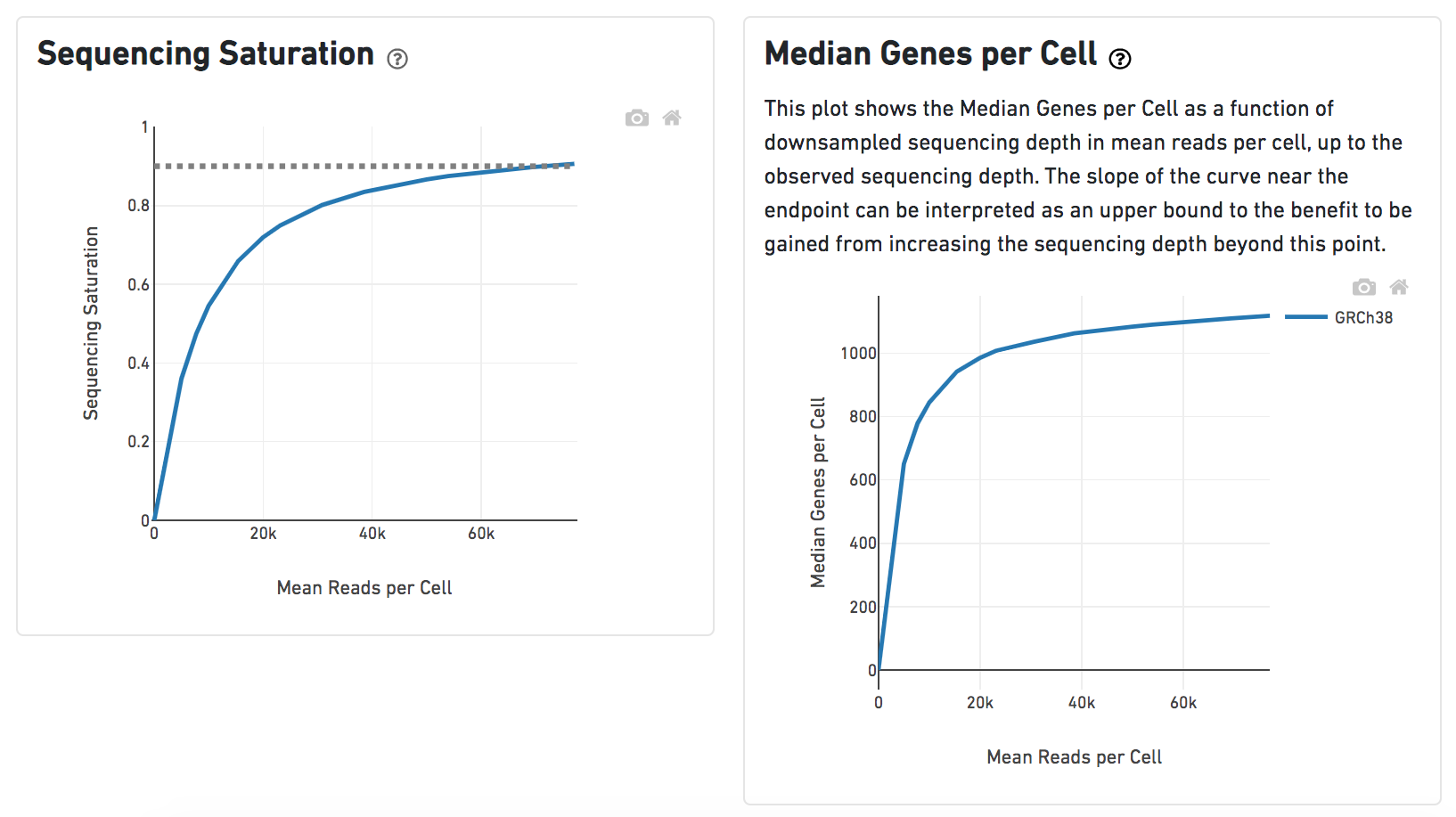
Other than two additional panels summarized information for Antibody Sequencing and Antibody Application have been added, the task report for Feature Barcode data is the same to scRNA-seq data report.
Users can click Configure to change the default settings In Advanced options (Figure 54).
Include introns: Count reads mapping to intronic regions. This may improve sensitivity for samples with a significant amount of pre-mRNA molecules, such as nuclei.
Expected cells: Expected number of recovered cells. Default: 3,000 cells.
Force cells: Force pipeline to use this number of cells, bypassing the cell detection algorithm. Use this if the number of cells estimated by Cell Ranger is not consistent with the barcode rank plot.
Memory limit (GB): Restricts Cell Ranger - Gene Expression to use specified amount of memory (in GB) to execute pipeline stages.
References
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/6.0/release-notes
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/4.0/release-notes
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/feature-bc-analysis#feature-ref
...