| Table of Contents |
|---|
| maxLevel | 2 |
|---|
| minLevel | 2 |
|---|
| exclude | Additional Assistance |
|---|
|
Classification in Partek Flow
Garnett[1] automated cell type classification has been wrapped as the Classification task in Partek Flow. Just like the original Garnett tool, Classification in Flow works on single-cell data, along with a cell type definition (marker) file, and trains a regression-based classifier. Once a classifier is obtained and published, it can be applied to classify future datasets from similar tissues. To improve the user experience, both maker file(.txt) and classifier file(.rds) have been implemented as library files in Flow.
Garnett classifiers in Flow
Flow hosts a few pre-trained classifiers as Managed classifiers. The list of available classifiers can be found here[2]. If a managed classifier exists for your data type, we recommend you try it. Besides the Managed classifiers, the Project classifiers trained on the same dataset prior to your classification can also be used to classify cell type. Project classifiers can be promoted to Managed classifiers if users publish them.
Classify cell type task in Flow
To classify cell type using a pre-trained classifier:
Select any non-normalized single cell data node, Filter counts here which contains raw count, choose Classify cell type task in Classification section (Figure 1)
| Numbered figure captions |
|---|
| SubtitleText | Selecting the Classify cell type task. |
|---|
| AnchorName | Task selection |
|---|
|
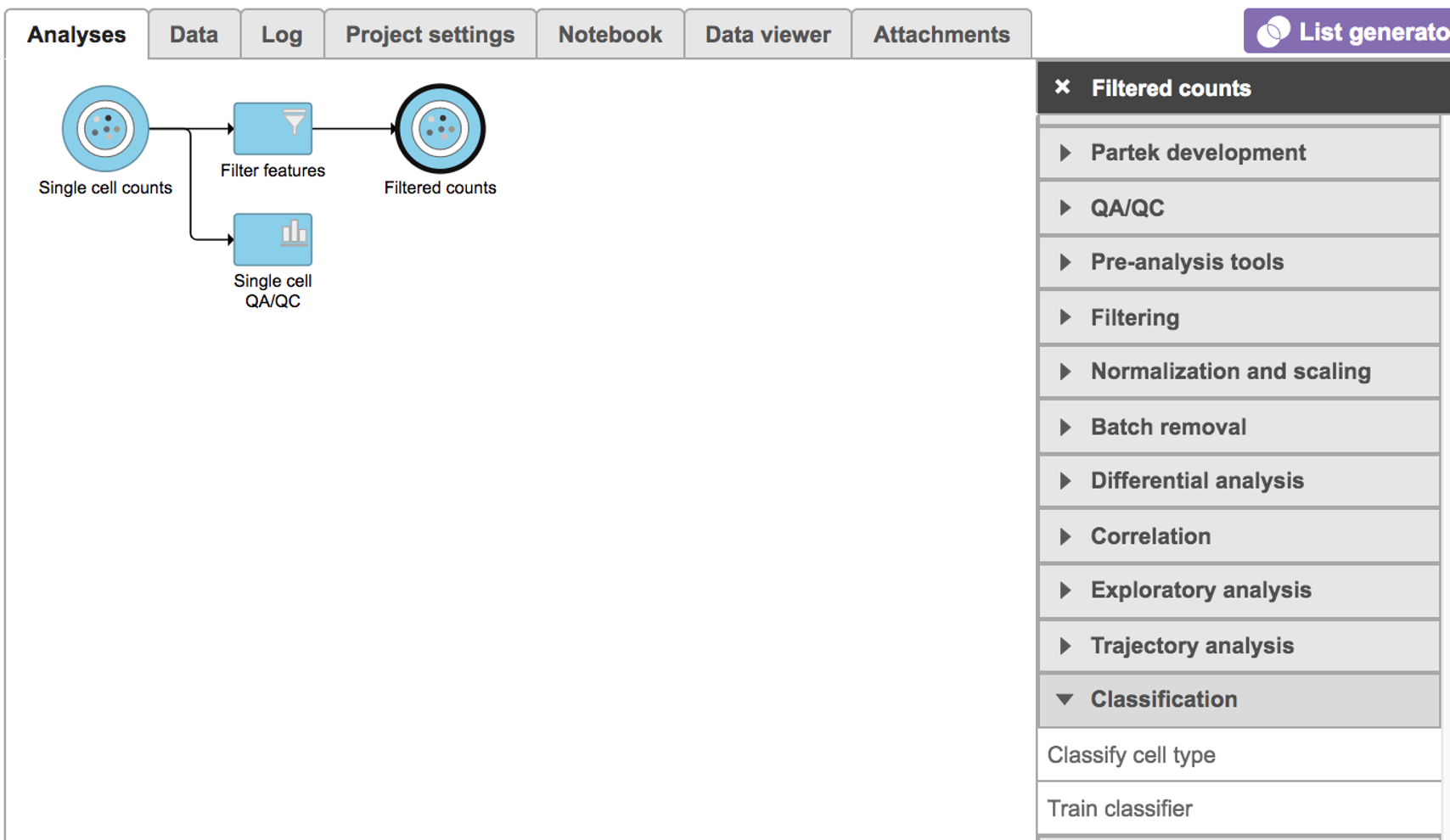
|
Managed classifiers users will be asked to create a new classifier file if it is the first time to run the task in Flow (Figure 2a). Users could select either Download Garnett classifier that matches the species and tissue type with the dataset working on from Flow maintaining list or Import Garnett classifier that’s trained out of Flow (Figure 2b). Then push the Create button to create the classifier file. Once the right classifier file has been created, Select Finish to start running the task (Figure 2d).
| Numbered figure captions |
|---|
| SubtitleText | Create Garnett classifier from Managed classifiers and run Classify cell type task with the selected classifier. |
|---|
| AnchorName | Task selection |
|---|
|
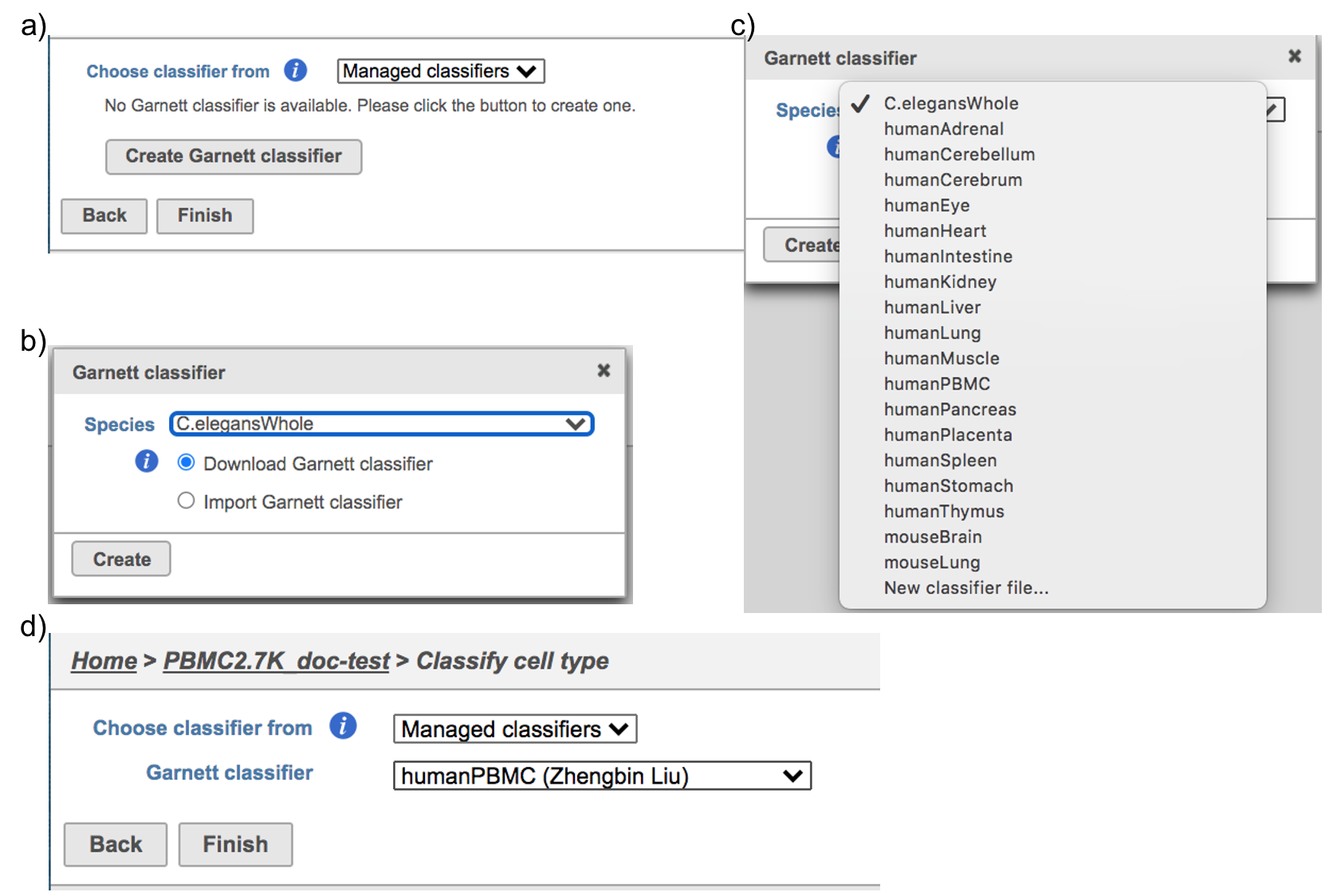
|
If users would like to classify cell type with the classifier obtained from the same project, ‘Project classifiers’ needs to be picked up from the dropdown list, then use ‘Select data node’ to choose the classifier before clicking the Finish button (Figure 3).
| Numbered figure captions |
|---|
| SubtitleText | Run Classify cell type task with Project classifier. |
|---|
| AnchorName | Task selection |
|---|
|
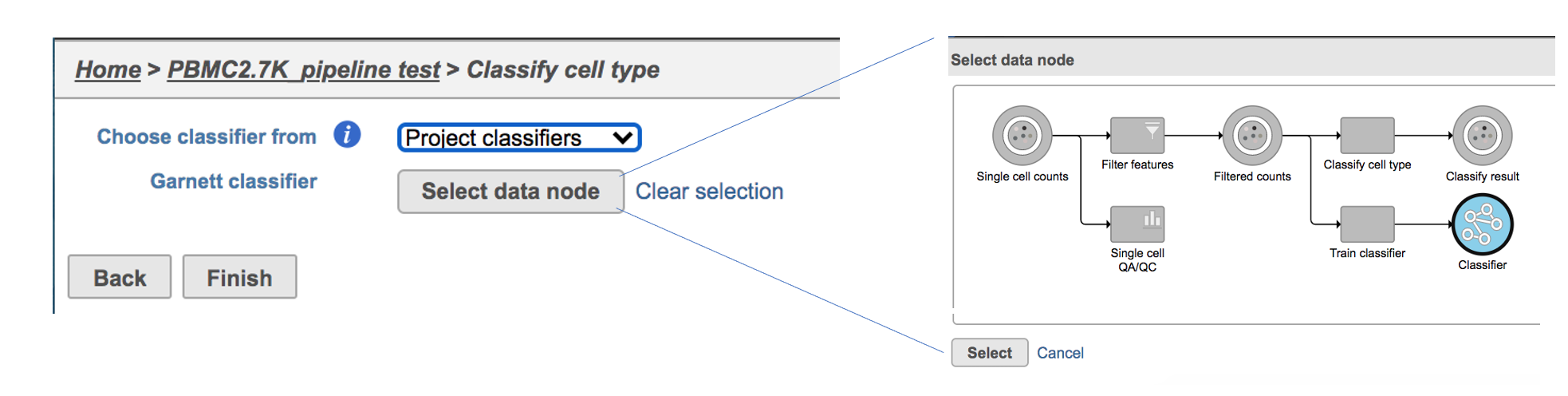
|
No matter which type of classifier was applied, it will output a new data node named Classify result (Figure 4). The outputs of cell type annotation are exactly the same as Garnett[1]. Downstream analysis like normalization, PCA, etc can be run on Classify result data node.
| Numbered figure captions |
|---|
| SubtitleText | Classify cell type task outputs a new data node. |
|---|
| AnchorName | Task selection |
|---|
|
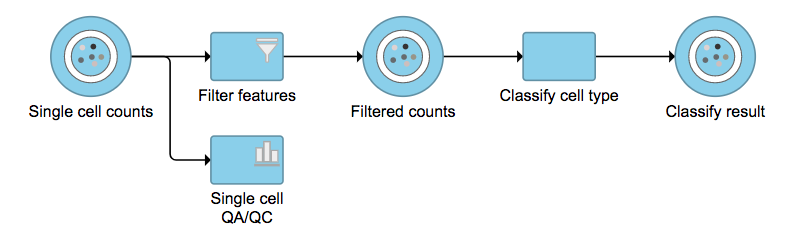
|
Train classifier in Flow
To train a classifier with a list of biomarkers with your own dataset:
Select any non-normalized single cell data node, Filter counts here, choose Train classifier task in Classification section (Figure 1)
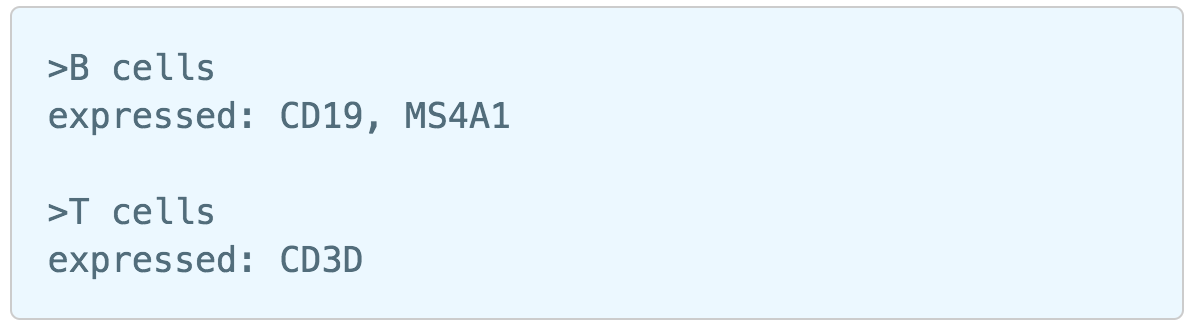
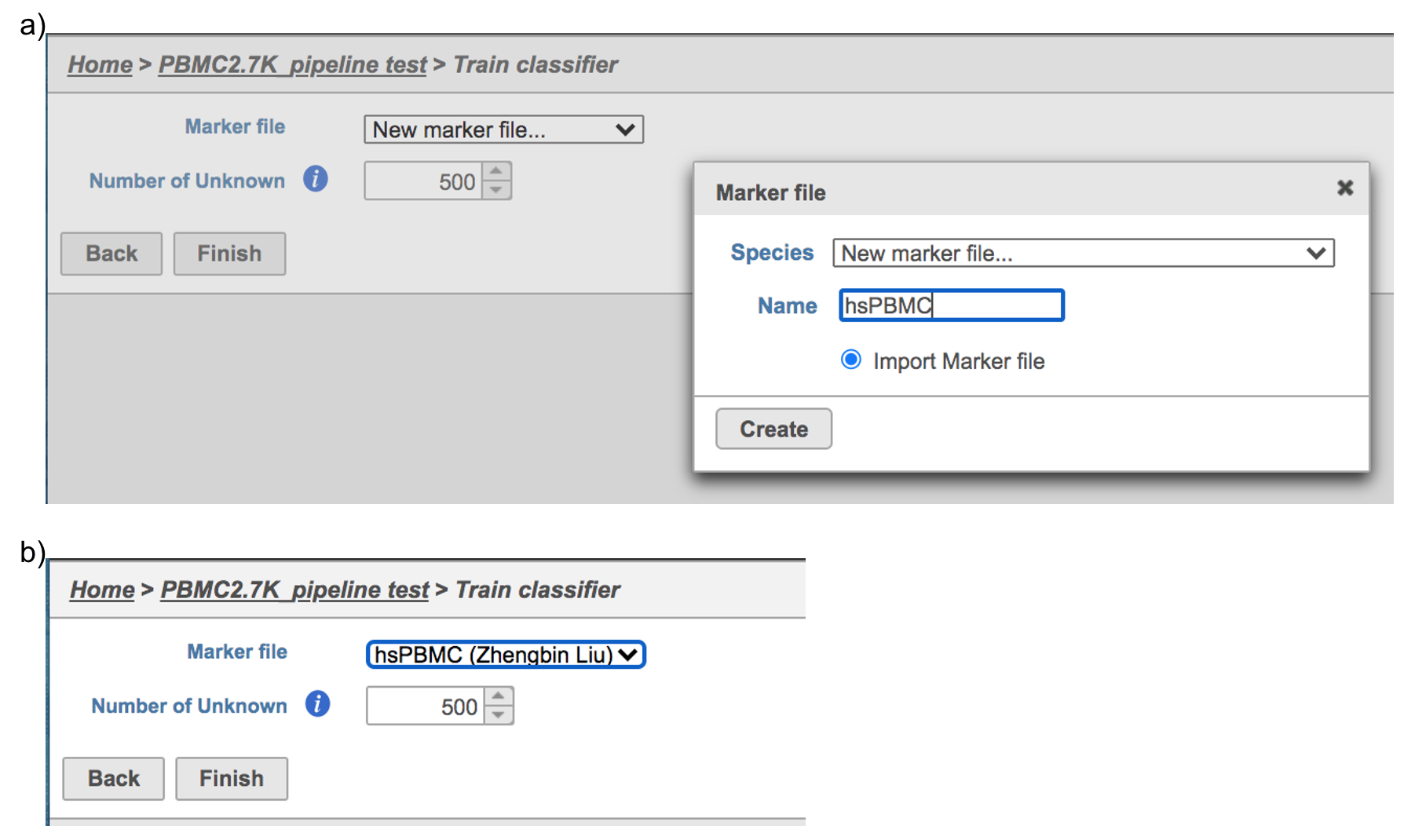
Similarly, first time users will be asked to create the Marker file . Partek Flow currently doesn’t host any marker files, users can add them as library files. Marker file should be a .txt file with the marker information in correct format. The same example in the Garnett tutorial of a simple valid Marker file is provided here (Figure 5). For more details on how to construct a Marker file, please refer to Garnett tutorial[3]. Click Create button, Flow will then save the file with the name that users provided for future use (Figure 6a).
| Numbered figure captions |
|---|
| SubtitleText | Example of marker file (.txt) from Garnett tutorial. |
|---|
| AnchorName | Task selection |
|---|
|
|
After the proper Marker file has been chosen, click the Finish button to start running the task as default (Figure 6b).
| Numbered figure captions |
|---|
| SubtitleText | Train Classifier task in Flow. |
|---|
| AnchorName | Task selection |
|---|
|
|
Train classifier task report in Flow
Once the task has been finished, click the Classifier datanode and choose the Task report in the Task results section, or prompt it to be one of the Managed classifiers by clicking the Publish classifier task in Classification section (Figure 7).
| Numbered figure captions |
|---|
| SubtitleText | Train classifier task report in Task results section. |
|---|
| AnchorName | Task selection |
|---|
|
|
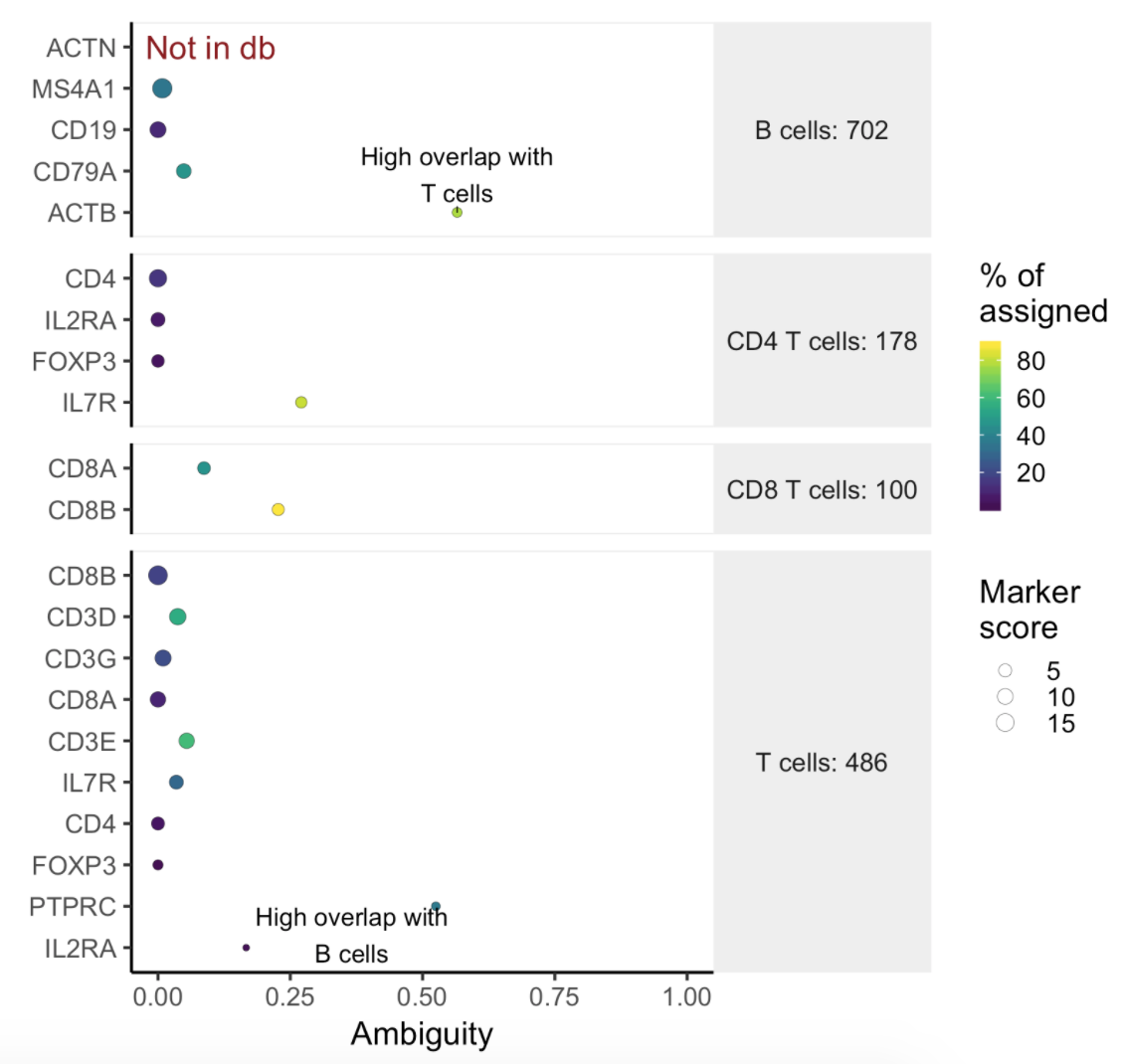
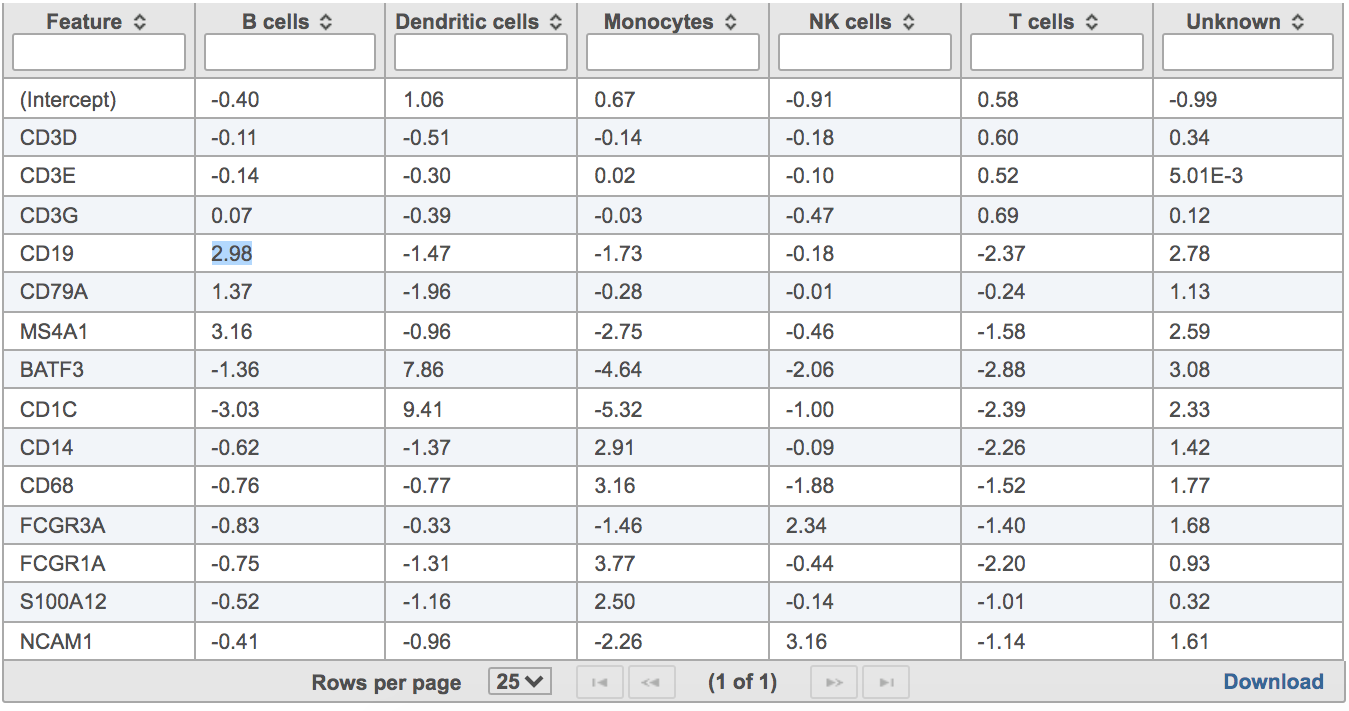
Users will find two parts in the task report: marker evaluation plot (Figure 8) and the classification gene table (Figure 9). The marker evaluation plot provides some key information about whether the chosen markers are good. Ambiguity scores are calculated for each of the markers which indicates how many cells receive ambiguous labels when this marker is included. The classification gene table could give a hint to which genes are chosen as the relevant genes for distinguishing between different cell types.
| Numbered figure captions |
|---|
| SubtitleText | Example of biomarker evaluation plot in task report. |
|---|
| AnchorName | Task selection |
|---|
|
|
| Numbered figure captions |
|---|
| SubtitleText | Example of classification gene table in task report. |
|---|
| AnchorName | Task selection |
|---|
|
|
Other parameters in this task that you can adjust include:
Number of Unknown: it tells Garnett how many outgroup cells it should compare against. The default is 500. For a dataset with fewer cells, the number should be smaller.
References
https://www.nature.com/articles/s41592-019-0535-3
https://cole-trapnell-lab.github.io/garnett/classifiers/
- https://cole-trapnell-lab.github.io/garnett/docs/#constructing-a-marker-file
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.








