...
To access the Partek on-line repository of user guides, tutorials, and whitepapers, go to your avatar and select Help. A new tab will be opened in your browser, showing the home page of the repository (Figure 1).
| Numbered figure captions |
|---|
| SubtitleText | Online repository of user guides, tutorials, and whitepapers |
|---|
| AnchorName | documentation.partek.com |
|---|
|
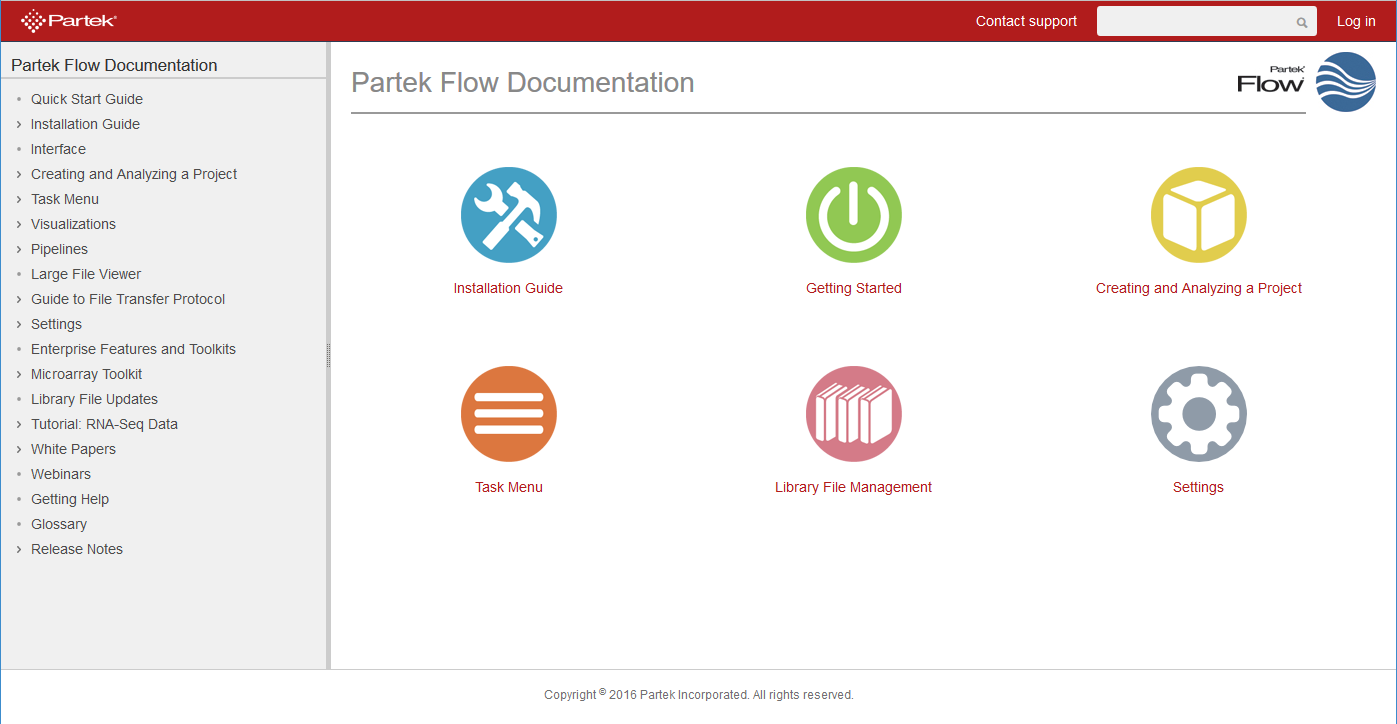 Image Modified Image Modified
|
Sample Data Sets
Two sample data sets can be downloaded directly to Partek Flow and you can use them to get familiar with the Partek Flow interface, tools, and pipelines. The download links are on the System information page (you can open it by selecting your avatar and then selecting the Settings option) (Figure 2).
| Numbered figure captions |
|---|
| SubtitleText | Downloading sample data sets |
|---|
| AnchorName | tutorial-data |
|---|
|
 Image Modified Image Modified
|
Depending on your preference, choose one of the sets. A new project will be automatically created (iDEA Challenge or Brain vs UHR) and Partek Flow will take you to a Data tab of that project (Figure 3). Since this is a tutorial project, there is no need to click on Import data as it will be done automatically.
| Numbered figure captions |
|---|
| SubtitleText | Creating a new project based on a sample data set. The project will be populated with samples automatically |
|---|
| AnchorName | new-project |
|---|
|
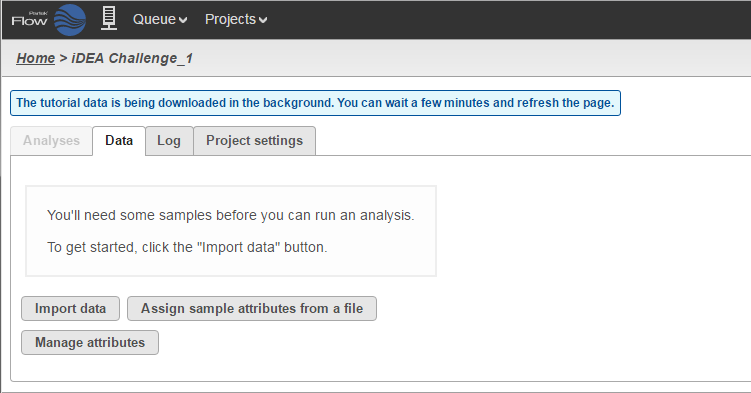 Image Modified Image Modified
|
At first the project is empty, but the file download will start automatically in the background. You can wait a few minutes and refresh the page or you can monitor the download progress in the Queue (Figure 4). You can access the Queue from the menu at the top of the page.
| Numbered figure captions |
|---|
| SubtitleText | Downloading tutorial data task in the queue (note that any user can download sample data set) |
|---|
| AnchorName | tutorial-download-task |
|---|
|
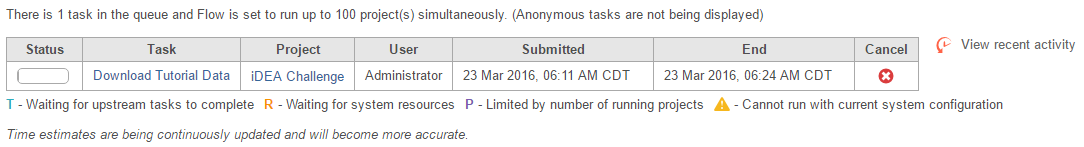 Image Modified Image Modified
|
iDEA Challenge project is based on RNA-seq experiment performed using Illumina paired-end reads. The samples are eight commonly used breast cancer cell lines; therefore, the sample set consists of 16 fastq files (total size of 790 MB). For downstream steps you may want to divide the samples into two groups, based on expression of estrogen receptor (ER). ER-positive cell lines are: BT474, MCF7, T47D, and ZR751; the remaining cell lines are ER-negative.
Brain vs. UHR project is also an RNA-seq experiment, but based on Ion Torrent sequencing (single-end chemistry). Two sets of samples are included: human brain samples (HBR) and universal human reference (UHR) samples. The project consists of 10 fastq files with the combined size of 1.4 GB.
...
Hovering over a task in the context sensitive menu (Figure 5) or the information icon  in the different task dialog (Figure 6) brings up a balloon with short description of the task / option.
in the different task dialog (Figure 6) brings up a balloon with short description of the task / option.
| Numbered figure captions |
|---|
| SubtitleText | Hover over a task in the context sensitive menu to get a balloon with a short description |
|---|
| AnchorName | toolbox-tooltip |
|---|
|
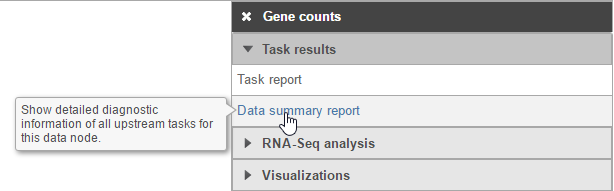 Image Modified Image Modified
|
| Numbered figure captions |
|---|
| SubtitleText | Hover over a tooltip icon to get a balloon with a short description |
|---|
| AnchorName | i-icon-tooltip |
|---|
|
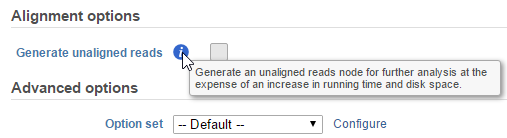 Image Modified Image Modified
|
...
Reporting a Problem
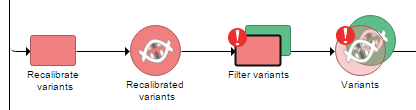
In an unlikely case of an error occurring during the analysis, the failed task will be highlighted by an exclamation mark (Figure 7). However, if the task does not fail altogether (there is no exclamation mark, and the task report is produced), the results can still me missing or questionable for some features. In that case, the issue might be resolved by following the directions in Differential Analysis Troubleshooting section.
| Numbered figure captions |
|---|
| SubtitleText | Failed task (in this example: Filter variants) highlighted by an exclamation mark. Note the the downstream task, i.e. Variants data node, is depicted as translucent to indicate that no result was produced |
|---|
| AnchorName | failed-task |
|---|
|
|
The In case of failure, the task node’s Task details page will contain Warning information section (optional) and the Error message section (Figure 8). In addition, you can download log files to your local computer (Download logs) or directly send them to Partek’s technical support team (Send logs to Partek), who will get back to you shortly.
| Numbered figure captions |
|---|
| SubtitleText | Task details page of a failed task. Warning information section is optional, the content of the Error message section depends on the type of error. Send logs to Partek sends the log files to Partek’s technical support team |
|---|
| AnchorName | task-details-error |
|---|
|
 Image Modified Image Modified
|
Using the flowstatus.sh diagnostic script
...
Phone numbers and support hours are also presented in the page if you wish to call us directly. There are also instructions on how to share screens if the support agent requests you during an online support call.
|
...




