Page History
Introduction
Gene-specific analysis (GSA) is a statistical modeling approach used to test for differential expression of genes or transcripts in Partek® Flow®. The terms "gene", "transcript" and "feature" are used interchangeably below. The goal of GSA is to identify a statistical model that is the best for a specific transcript, and then use the best model to test for differential expression.
Overview of GSA methodology
To test for differential expression, a typical approach is to pick a certain parametric model that can deliver p-values for the factors of interest. Each parametric model is defined by two components:
...
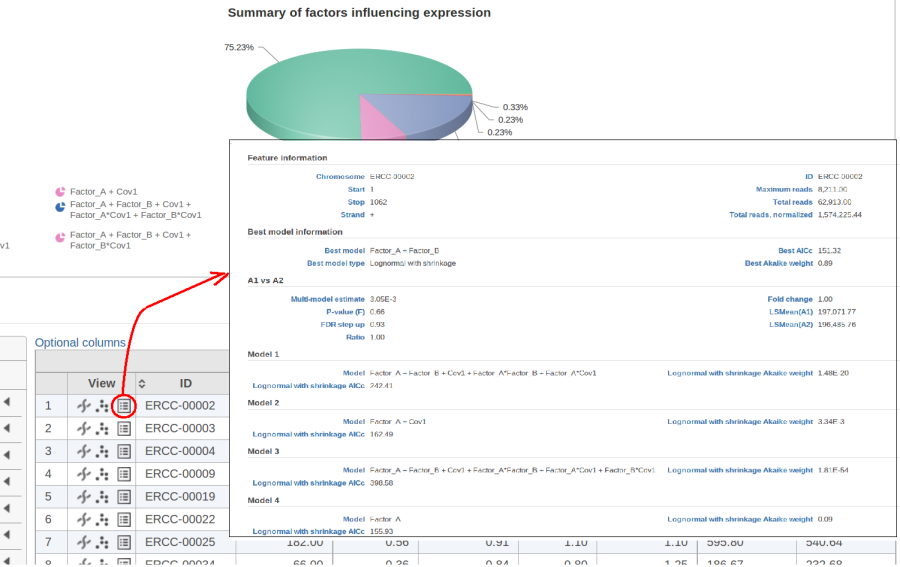
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Obtaining reproducible results in GSA
GSA is a great tool for automatic model selection: the user does not have to spend time and effort on picking the best model by hand. In addition, GSA allows one to run studies with no replication (one sample per condition) by specifying Poisson distribution in Advanced Options (Figure 1). However, not having enough replication is likely to deliver the results that are far more descriptive of the dataset at hand rather than the population from which the dataset was sampled. For instance, in the example from the previous section the number of fitted models (15) exceeds the number of observations (12), and therefore the results are unlikely to be representative of the population. If the study were to be reproduced with some 12 new samples, the list of significant features could be very different.
...
Second, the error degrees of freedom threshold in Default mode is adjusted automatically for the purpose of excluding designs that have too many terms compared to the number of observations. Third, the hierarchical design restriction explained in the previous section is also aimed at keeping the number of considered designs reasonable. The user can override the first two restrictions by switching from Default to Custom mode in Advanced Options (Figure 1). The 2nd order and hierarchical design restrictions cannot be disabled in GSA but they are absent in Flow ANOVA. The latter is similar to "Lognormal" option in GSA.
Using shrinkage plots
When "Lognormal with shrinkage" is enabled, a separate shrinkage plot is displayed for each design (Figure 4). First, a lognormal linear model is fitted for each gene separately, and the standard deviations of residual errors are obtained (green dots in the plot). Applying shrinkage amounts to two more steps. We look at how the errors change depending on the average gene expression and we estimate the corresponding trend (black curve). Finally, the original error terms are adjusted (shrunk) towards the trend (red dots). The adjusted error terms are plugged back into the lognormal model to obtain the reported results such as p-value.
...
Using multimodel inference appears to be a better alternative to the ad hoc method in DESeq2 that switches shrinkage on and off all the way. Once again, it is both technically possible and emotionally tempting to automate the handling of abnormal features by enabling both Lognormal models in GSA and applying them to all of the transcripts. Unfortunately, that can make the results less reproducible overall, even though it is likely to yield more accurate conclusions about the drastically outlying features.
References
[1] Auer, 2011, A two-stage Poisson model for testing RNA-Seq.
[2] Burnham, Anderson, 2010, Model selection and multimodel inference.
[3] Dillies et al, 2012, A comprehensive evaluation of normalization methods.
[4] Storey, 2002, A direct approach to false discovery rates.
[5] Law et al, 2014, Voom: precision weights unlock linear model analysis. Note that this paper introduces both "limma trend" and "limma voom", but the present implementation in GSA corresponds to "limma trend".
[6] Love et al, 2014, Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2.
[7] Bioconductor support forum. Accessed last: 4/12/16. https://support.bioconductor.org/p/80745/#80758
...