Join us for a webinar: The complexities of spatial multiomics unraveled
May 2
Page History
| Table of Contents | ||
|---|---|---|
|
What is Gene set enrichment?
...
Running Gene set enrichment
We recommend filtering to a set of genes you want to test for enrichment, but Gene set enrichment will run on any Feature list data node.
- Click a Feature list data node
- Click the Biological interpretation section of the toolbox
- Click Gene set enrichment
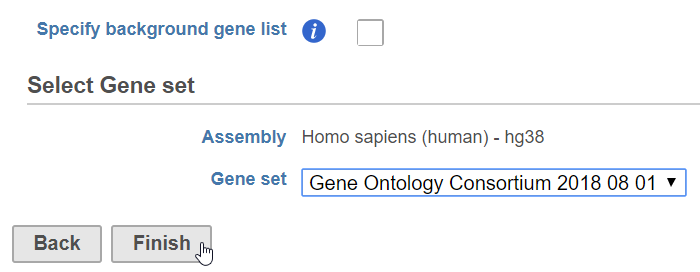
- Configure the background gene list (optional)
The background gene list is used as the list of possible genes. By default, this is the genes included in the selected gene set database. If your assay limits the genes that could be detected, you may want to specify a background list.
- Choose the Gene set
The gene sets available for the current Assembly are listed under the Gene set drop-down list. The assembly is automatically selected, if possible. If the assembly cannot be detected, you can specify it using a drop-down menu.
- Click Finish to run (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Overview
Content Tools