Page History
...
Cell Ranger is a set of analysis pipelines that process Chromium single-cell RNA-seq output to align reads, generate feature-barcode matrices and perform clustering and gene expression analysis for 10X Genomics Chromium Technology[1].
Cell ranger in Partek Flow
Cell Ranger v6.0.0[2] has been wrapped In Partek® Flow® as Cell ranger task. It does not comprehensively cover all of the options and analysis cases Cell Ranger can handle for now, but converts FASTQ files from cellranger mkfastq and performs alignment, filtering, barcode counting, UMI counting. The output gene expression count matrix in .h5 format (both raw and filtered available for users to download in the output page of task details) then becomes the starting point for downstream analysis for scRNA-seq in Flow.
...
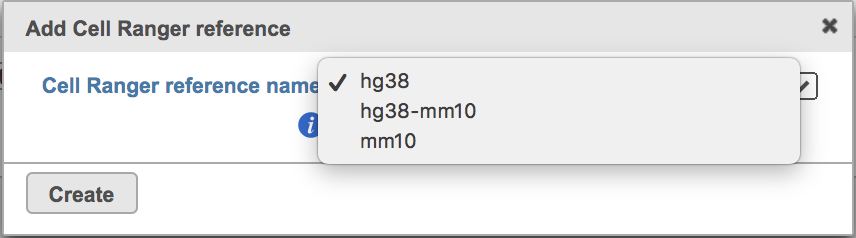
Clicking the big grey button of Create Cell Ranger 6.0.0 reference would pop up a new window where lists the three pre-built reference genomes for human(hg38), mouse(mm10) and the mix of two(hg38-mm10), respectively (Figure 3). They are exactly the same reference genomes (2020-A) that are provided in Cell Ranger by default. In details, the transcriptome annotations are respectively GENCODE v32 for human and vM23 for mouse, which are equivalent to Ensembl 98[3]. References for other organisms currently are not available in Flow Cell ranger, and will be coming in the next versionfuture.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
A new data node named ‘Single cell counts’ will be displayed in Flow if the task is running (Figure 6). To This data node contains filtered feature barcode count matrix. To open the task report when the task is finished, simply double click the output data node, or select the ‘Task report’ in the section after single clicking the data node once the tasking running was finished. Users then will find the task report (Figure 7) is the same to the ‘Summary HTML’ from Cell Ranger output.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
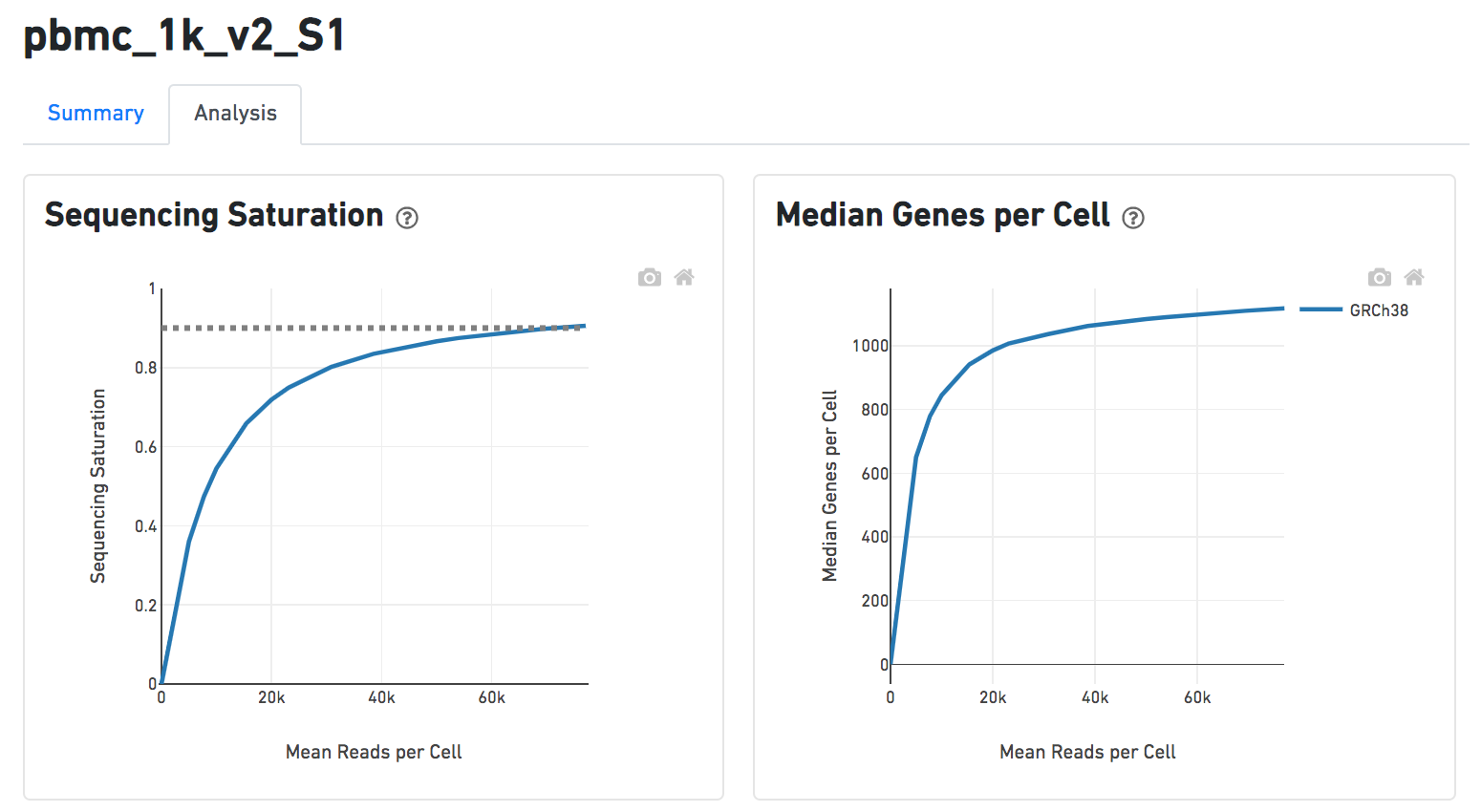
A lot of details will Details will be exhibited and the panel will be expanded correspondingly if the the symbolin the panel of each report has been clicked icon is clicked. In the example below, the plot of Median Genes per Cell has been expanded while the Sequencing Saturation plot hasn't (Figure 9).
...