Page History
...
To perform pseudotime analysis, you need to point to the cells at the beginning of the biological process you are interested in. For example, cells at the earliest stage of differentiation sequence. There are two ways to perform pseudotime analysis in Partek Flow, depending on the way the root node nodes (=cells at the beginning of pseudotime) is are defined.
- Manual selection of root node. The user has to specify the root nodes (one or more).
- Automatic selection of the root node. The root node is nodes are picked by the algorithm.
Manual Selection of the Root Node
...
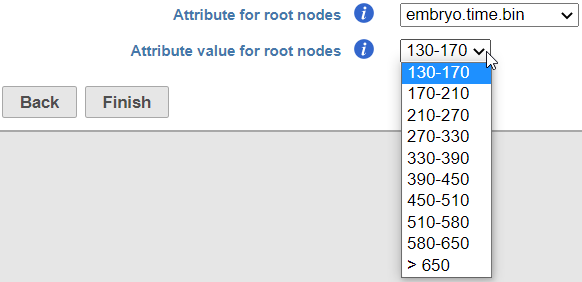
If suitable meta-data are available, it is possible to automatically select the root node. For example, you may know which cells were harvested from the earliest time points. The cells need to be annotated by that information (Annotate Cells task) before running Trajectory Analysis. The annotation will, in turn, be available in the Trajectory analysis setup dialog, upon selecting the Programmatically calculate default root nodes option (Figure xx).
- Attribute for root nodes. The drop down list will show the available cell-level attributes. Specify the one which should be used to identify the root nodes. In the example in Figure xx, the attribute embryo.time.bin describes the developmental time point at which the cells were harvested.
- Attribute value for root nodes. The drop down list will show the content of the attribute selected under Attribute for root nodes. Specify the one that corresponds to the earliest time point (i.e. beginning of pseuodtime). In the example in Figure xx, the earliest time point is the time bin 130-170.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
If selectedOnce the options have been set, Monocle 3 will first group the cells according to which trajectory node they are nearest to. It then calculates the fraction of the cells from the earliest time point at each trajectory node. Finally, it picks the node with the highest prevalence of the early cells and treats it as the root node.
In Figure xx, node #1 has been picked as the beginning of pseudotime, due to the high number of cells from the embryo.time.bin 130-170 grouped around that node (a 2D view is shown).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Difference Between Monocle 3 and Monocle 2
...