Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
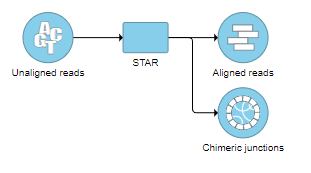
Selecting the Chimeric results node opens the toolbox task menu (Figure 13) with twooptions: Data summary report or Download data.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
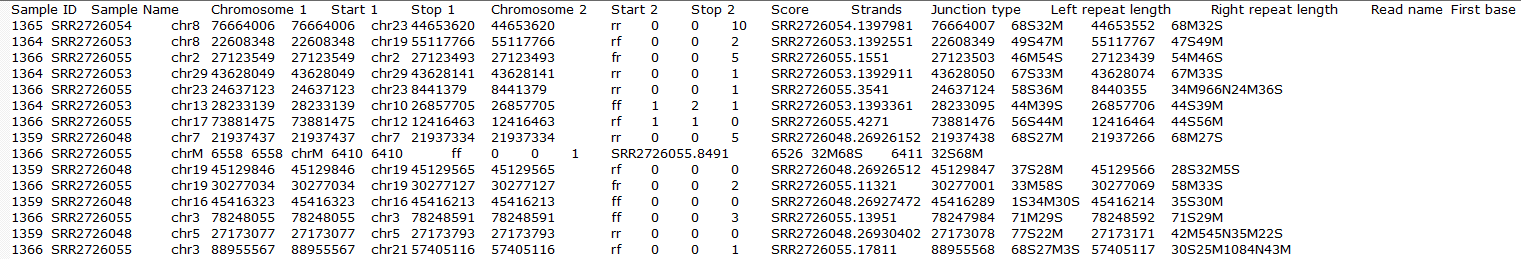
Clicking on the Download data results in download of a .fusion file to the local computer. The file is human readible readable and can be opened in a text editor (example in Figure 14). For details refer to STAR's documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
References
- Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192-200.
- Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134-142.
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011;12:R72
- TopHat-Fusion. An algorithm for discovery of novel fusion transcripts. http:// http://tophat.cbcb.umd.edu/fusion_index.html Accessed on April 25, 2014
- Dobin A, Davies CA, Schlesinger F et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21.
...
Overview
Content Tools