Page History
...
- Click the counts data node
- Click the Exploratory analysis section of the toolbox
- Click Graph-based clustering
- Configure the parameters
- Click Finish to run
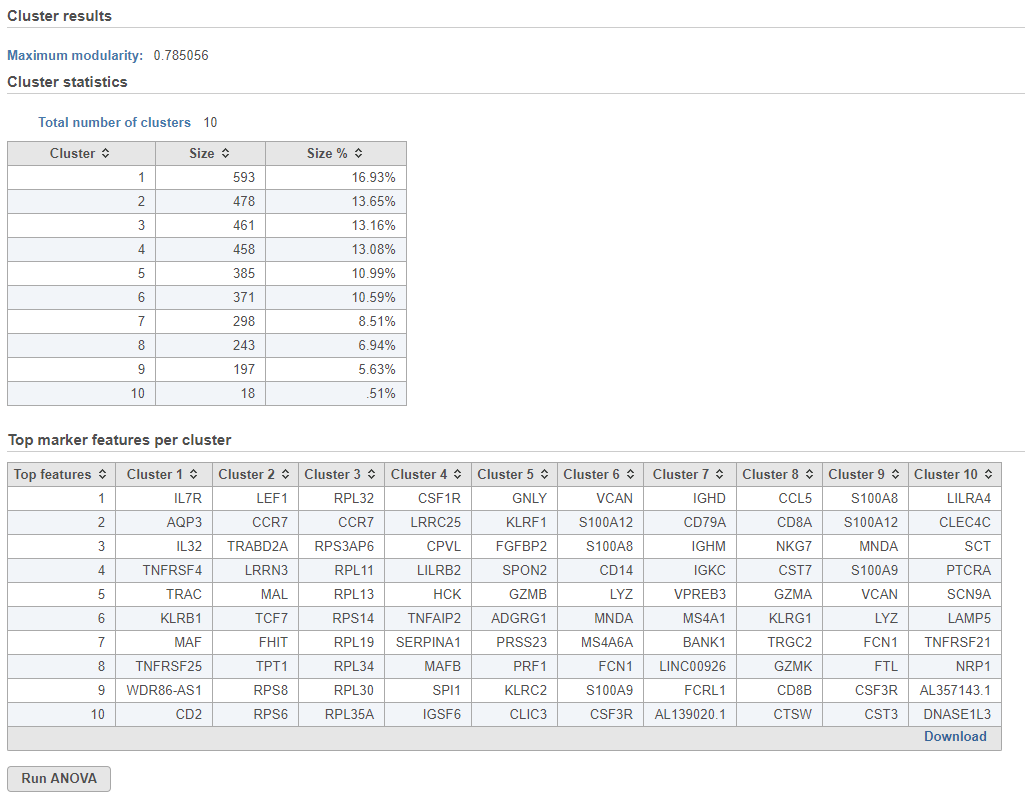
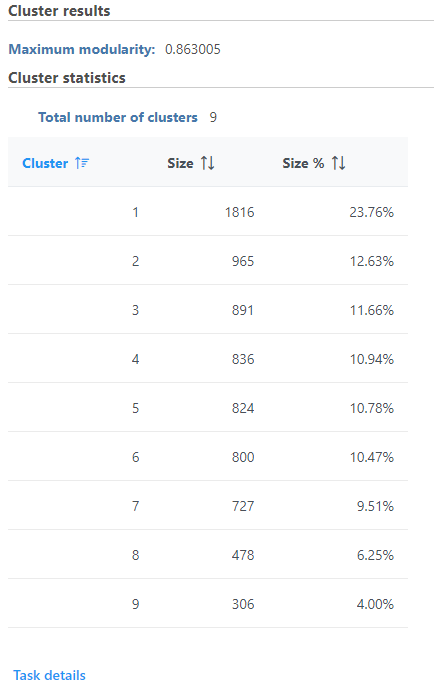
Graph-based clustering produces a Clustering result data node. The task report lists the cluster results , and cluster statistics , and top marker features per cluster (Figure 1). If clustering was run with Split cells by sample enabled on a single cell counts data node, the cluster results table displays the number of clusters found for each sample and clicking the sample name opens the sample-level report.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Cluster results
...
The total number of clusters is listed along with the number and percentage of cells in each cluster.
Top marker features per cluster
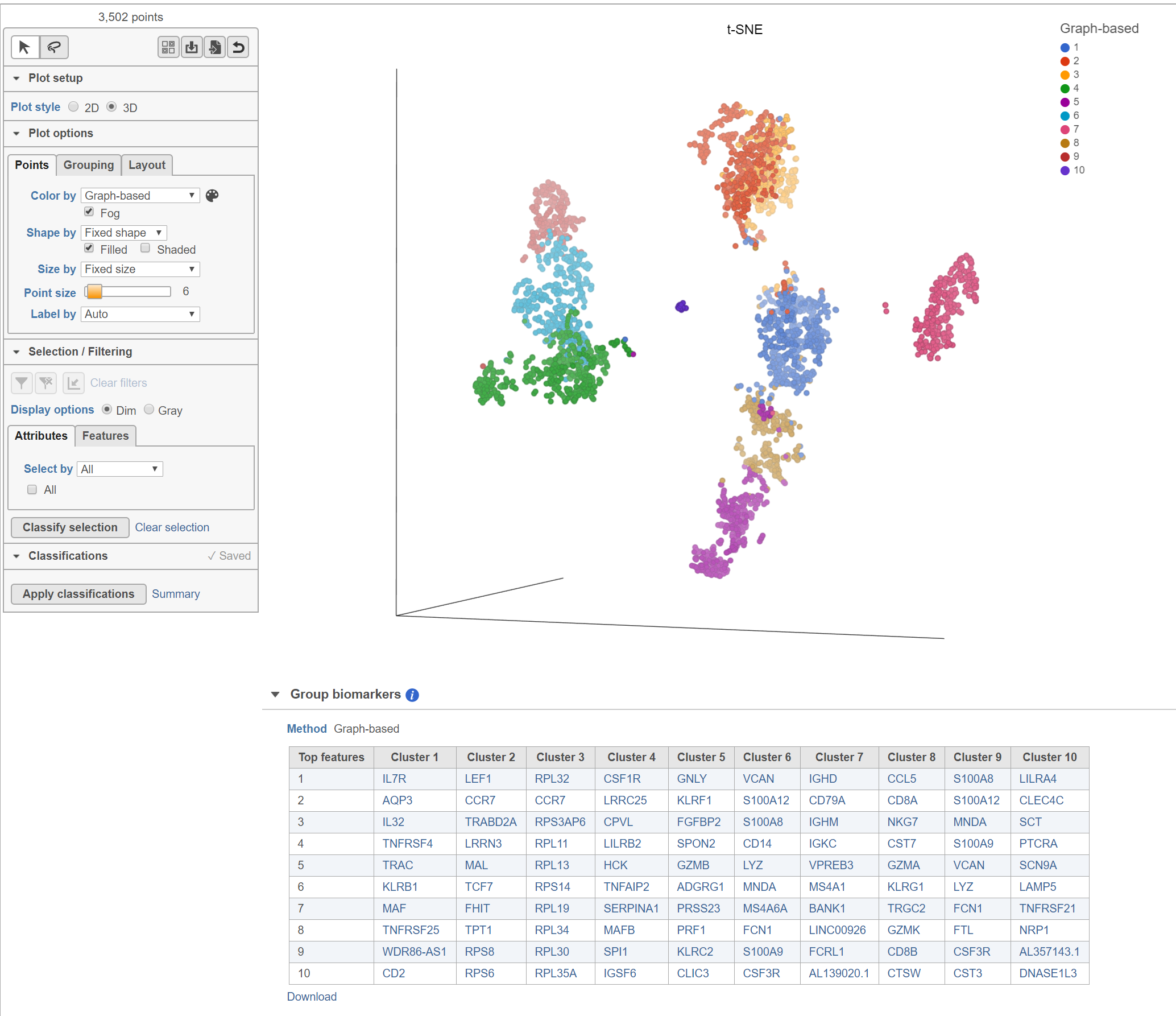
Biomarkers for each cluster are calculated using an ANVOA test where each cluster is compared to the other cells in the data set, genes with fold-change > 1.5 are included, and these genes are sorted by ascending p-value (ties broken by greater fold change). The top 10 genes for each cluster are shown in the table. The full ANOVA results can be obtained by clicking the Run ANOVA button, which will generate a Feature list data node.
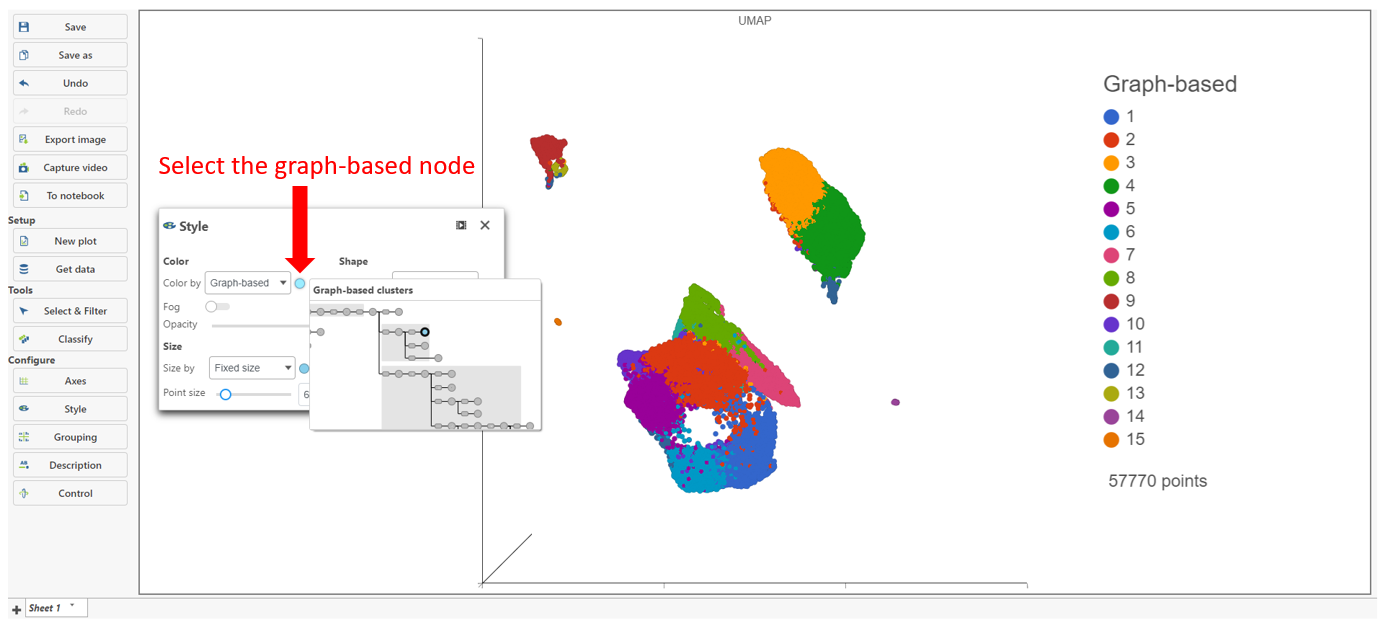
The Clustering result data node includes the input values for each gene and adds cluster assignment as a new attribute, Graph-based, for each observation. If the Clustering result data node is visualized by Scatter plot, PCA, t-SNE, or UMAP, the plot will be colored by the Graph-based attribute and the group biomarker table, if generated, will be included below the plot (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Basic Graph-based clustering parameters
...
Choose which version of the Louvain clustering algorithm to use. Options are Louvain [1], Louvain with refinement [2], and SLM [3]. The most recent version is Smart Local Moving (SLM). The default is Louvain.
Compute biomarkers will compute features that are highly expressed when comparing each cluster.
Split cells by sample
Chose whether to run Graph-based clustering on all samples together or on each sample individually.
...