Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
Scientists often develop lists of genes, probes, transcripts, SNPs, and genomic regions of interest from analysis tools, research papers, and databases. Using Partek®
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
Genomics Suite®, these lists can be integrated with genomics data sets, analyzed with powerful statistics, and visualized for new insights.
This tutorial will illustrate how to:
Importing a text file
The preferred method for importing a generic list of data into Partek Genomics Suite is as a text file.
- Select File from the main toolbar
- Select Text (.csv .txt)...under the Import option
- Select the text file to launch the Import .txt, .tsv, or .csv File dialog
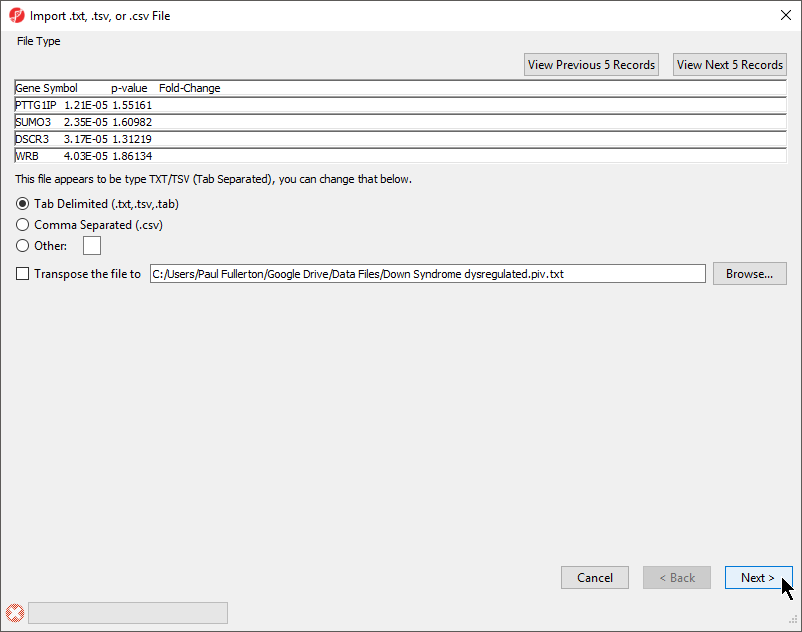
The File Type section of the Import dialog includes a preview of the text file and import options (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The columns in the import file can be separate by a tab, comma, or any other character.
For most applications, the items on the list should be in rows while attributes or values should be in columns. If a list is oriented with items on columns, select Transpose the file to to import a transposed spreadsheet.
- Select Next > to move to the Data Type section
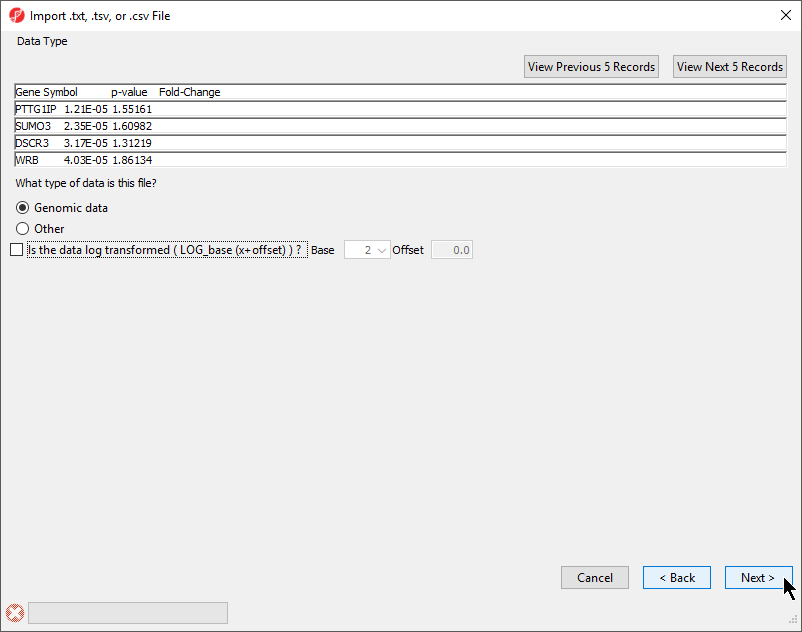
- Select your data type; here we have chosen Genomic Data because it is a gene list (Figure 2)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
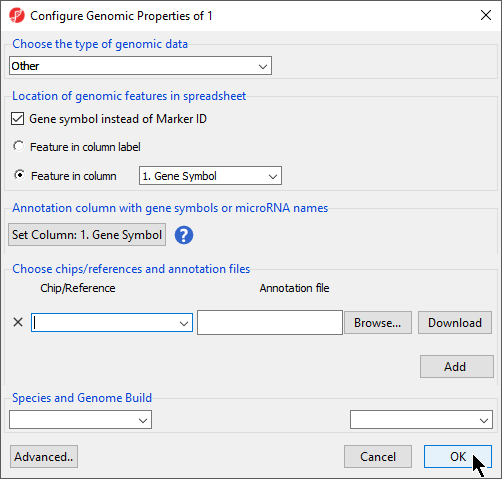
Selecting Genomic Data will result in a dialog prompt to configure genomic properties including selecting the type of genomic data, the location of genomic features in the spreadsheet, the annotation column with gene symbols, the chip or reference source and annotation file, and the species and genome build. This option should be selected if the text file contains genomic position data or other array/sequencing results.
- Select Next >
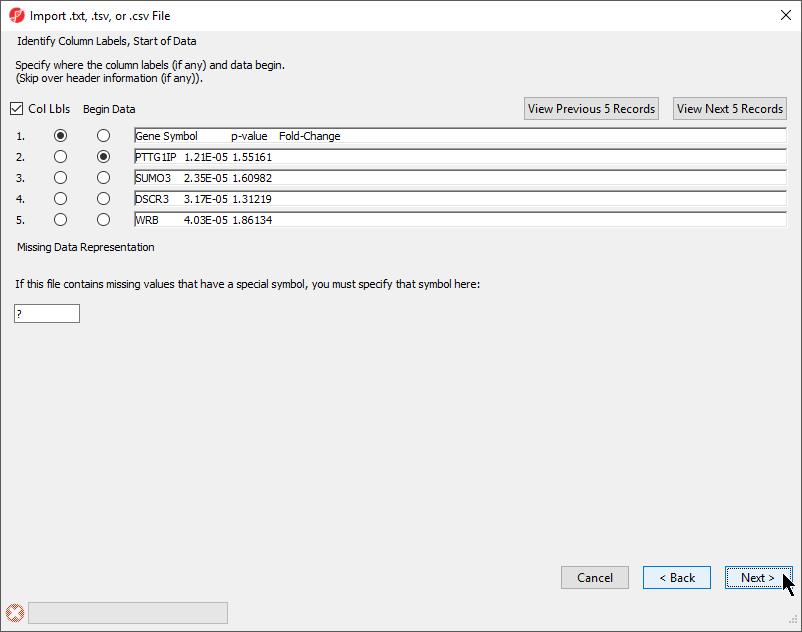
The Identify Column Labels, Start of Data section (Figure 3)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The next step is to identify where the data starts and where the optional header is found. The line that contains the header (if present) must precede the data. If there are lines to be skipped in the file (like comments), they may only appear at the top of the file, before the header line or data begin.
If there are many comment lines at the start of the file, you may need to select View Next 5 Records to get to the row that contains the column header. If you accidentally move past the screen that contains the header or data rows, select View Previous 5 Records.
If there are missing numerical values or empty cells in your input list, insert a special character or symbol (?, N/A, NA, etc.) in the missing cells; you will specify the character in the Missing Data Representation section of the dialog
- If a header row is present, select Col Lbls to allow you to select a column header row
- Select the row where the data beings using the Begin Data selector
- If any cells have a missing value, you can signify this with a special symbol selected using the Missing Data Representation panel
This is important if the missing value is a number in a column that you plan to use for statistical analysis. The default missing value indicator is ?.
- Select Next >
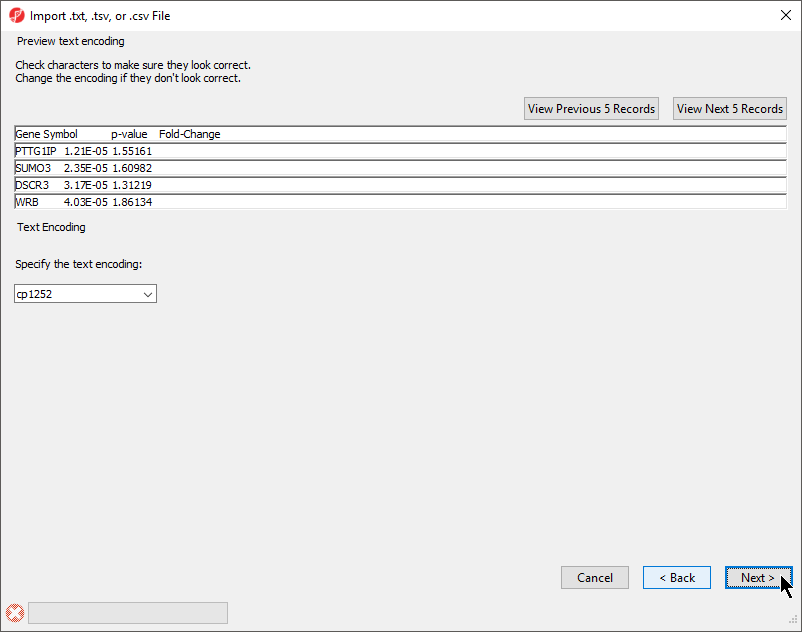
The Preview text encoding section (Figure 4) previews the first five lines of the file, allowing you to check if the text encoding is correct.
- If the text does not appear properly, use the Specify the text encoding: drop-down menu to choose the correct encoding
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Select Next >
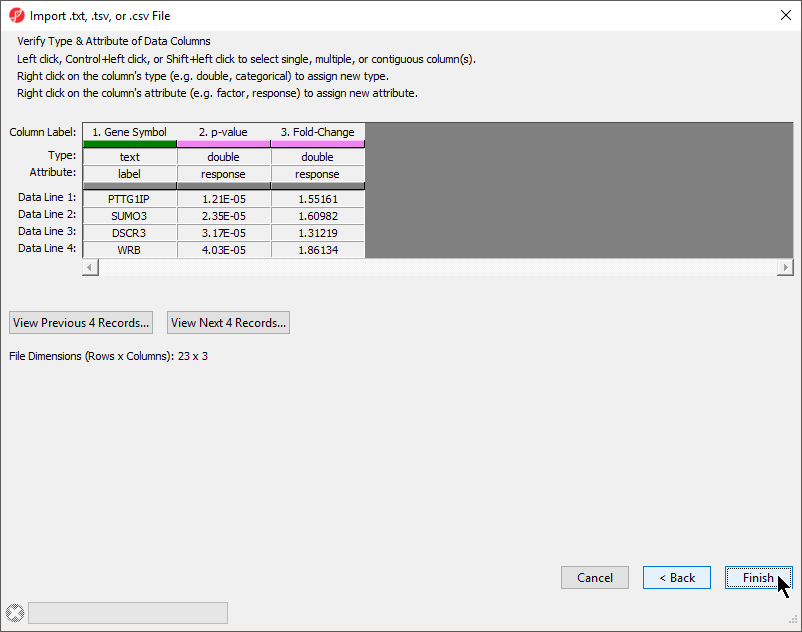
The final section of the Import .txt, .tsv, or .csv File dialog is Verify Type & Attribute of Data Columns (Figure 5). While data column type and attribute can be modified after import, it is easier and faster to select the proper options during import as multiple columns may be selected during this dialog.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Check and modify column types and attributes
If there is an identifier like gene symbol or SNP, the Type field for that column should be set to text and Attribute should be set to label. Numeric values (intensities, p-values, fold-changes, etc.) should have Type set to double and Attribute set to response. The other possible value for Attribute is factor and describes sample data. The user interface is this dialog allows you to select multiple columns at once. The interface controls are detailed in the dialog (Figure 5).
- Select Finish to import the text file and open it as a spreadsheet
If Genomic Data was selected in the Data Type section, the Configure Genomic Properties dialog will open (Figure 6)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|