Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
What is Hierarchical Clustering?
Hierarchical clustering is used to group groups similar objects into “clusters”. In the beginningclusters. To start, each row and/or column is considered a cluster. In hierarchical clustering, the The two most similar clusters are then combined and continue to combine this process is iterated until all objects are in the same cluster. Hierarchical clustering produces a tree (called a dendogram) that shows the Hierarchical clustering displays the resulting hierarchy of the clusters . This allows in a tree called a dendrogram. Hierarchical clustering is useful for exploratory analysis to see how the microarrays because it shows how samples group together based on similarity of features. Hierarchical
Hierarchical clustering is considered an unsupervised clustering method. Unsupervised Unsupervised clustering does methods do not take any of the the identity or attributes of samples into account when clustering. This means that experimental variables such as treatment, phenotype, tissue, etc. into account while clustering, whereas supervised clusters does number of expected groups, etc. do not guide or bias cluster building. Supervised clustering methods do consider experimental variables when building clusters.
Visualizing Hierarchical Clustering
To illustrate the capabilities and customization options of hierarchical clustering . Partek offers an alternative to Hierarchical clustering in the form of K-Means clustering and Self-Organizing Map. You can read a more in depth description of how Partek performs these different forms for clustering analysis in Chapter 8 Hierarchical & Partitioning Clustering of the Partek Manual. The Partek user’s manual is embedded in Partek Genomics Suite under Help > On-Line Help.
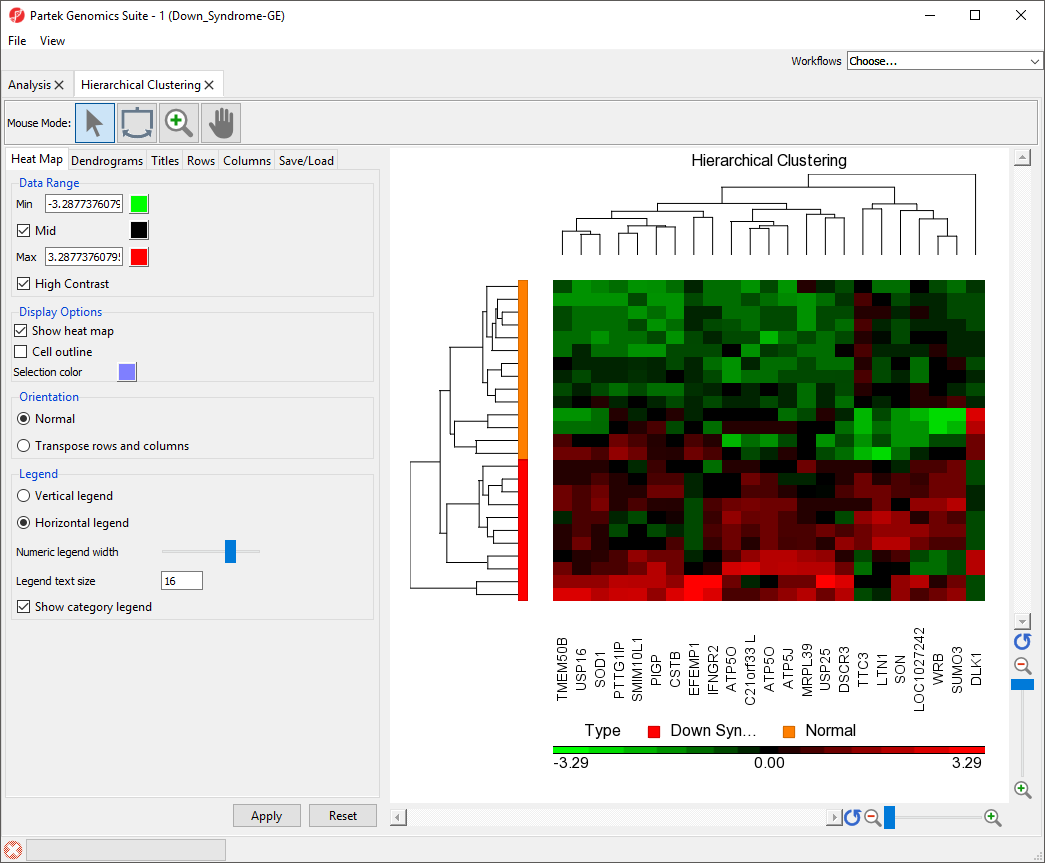
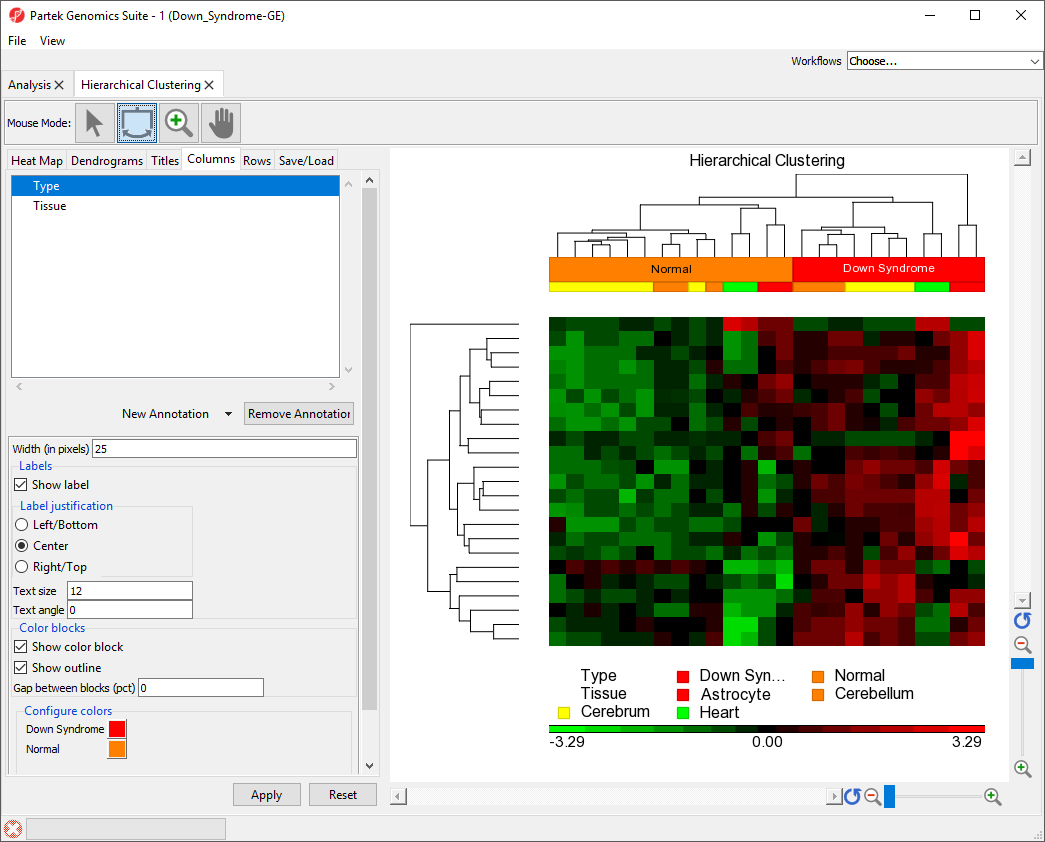
in Partek Genomics Suite, we will explore an example of hierarchical clustering drawn from the tutorial Gene Expression Analysis. The data set in this tutorial includes gene expression data from patients with or without Down syndrome. Using this data set, 23 highly differentially expressed genes between Down syndrome and normal patient tissues were identified. These 23 differentially regulated genes were then used to perform hierarchical clustering of the samples. Follow the steps outlined in Performing hierarchical clustering to perform hierarchical clustering and launch the Hierarchical Clustering tab (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The right-hand section of the Hierarchical Clustering tab is a heat map showing relative expression of the genes in the list used to perform clustering. The heat map can be configured using the properties panel on the left-hand side of the tab. In this example, the low expression value is colored in green, the high expression value is in red, and the mid-point value between min and max is colored in black.The dendrograms on the left-hand side and top of the heat map show clustering of samples as rows and features (probes/genes in this example) as columns. Columns are labeled with the gene symbol if there is enough space for every gene to be annotated. Rows are colored based on the groups of the first sample categorical attribute in the source spreadsheet. The sample legend below the heat map indicates which colors correspond to which attribute group. In this example, Down syndrome patient samples are red and normal patient samples are orange.
The heat map can be configured using the properties panel on the left-hand side of the Hierarchical clustering tab.
Configuring the Hierarchical Clustering Plot
| Table of Contents | ||||
|---|---|---|---|---|
|
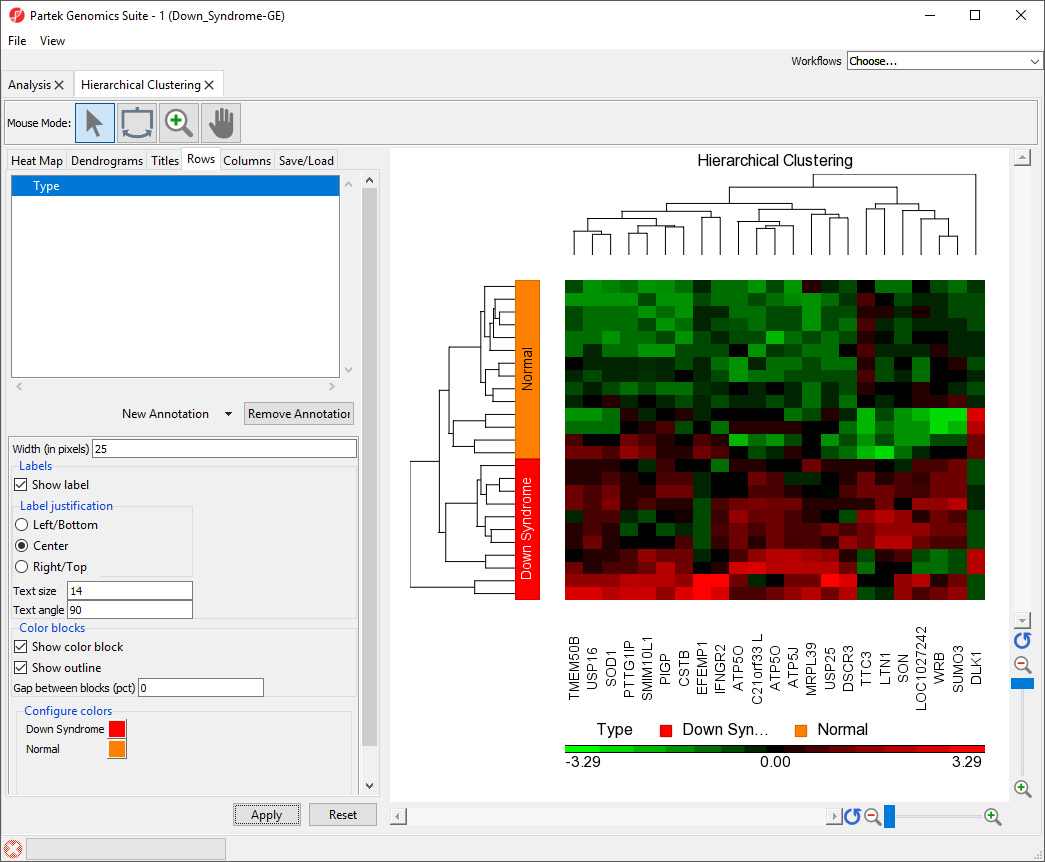
Labeling Sample Groups in the Heat Map
- Select the Rows tab
- Verify that Type appears in the annotation box
- Set Width (in pixels) to 25
This will increase the width of the color box indicating sample Type.
- Select Show Label
- Set Text size to 12
- Set Text angle to 90
This angle is relative to the x-axis. When set to 90, the text will run along the y-axis.
- Select Apply
The sample attributes are now labeled with group titles (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
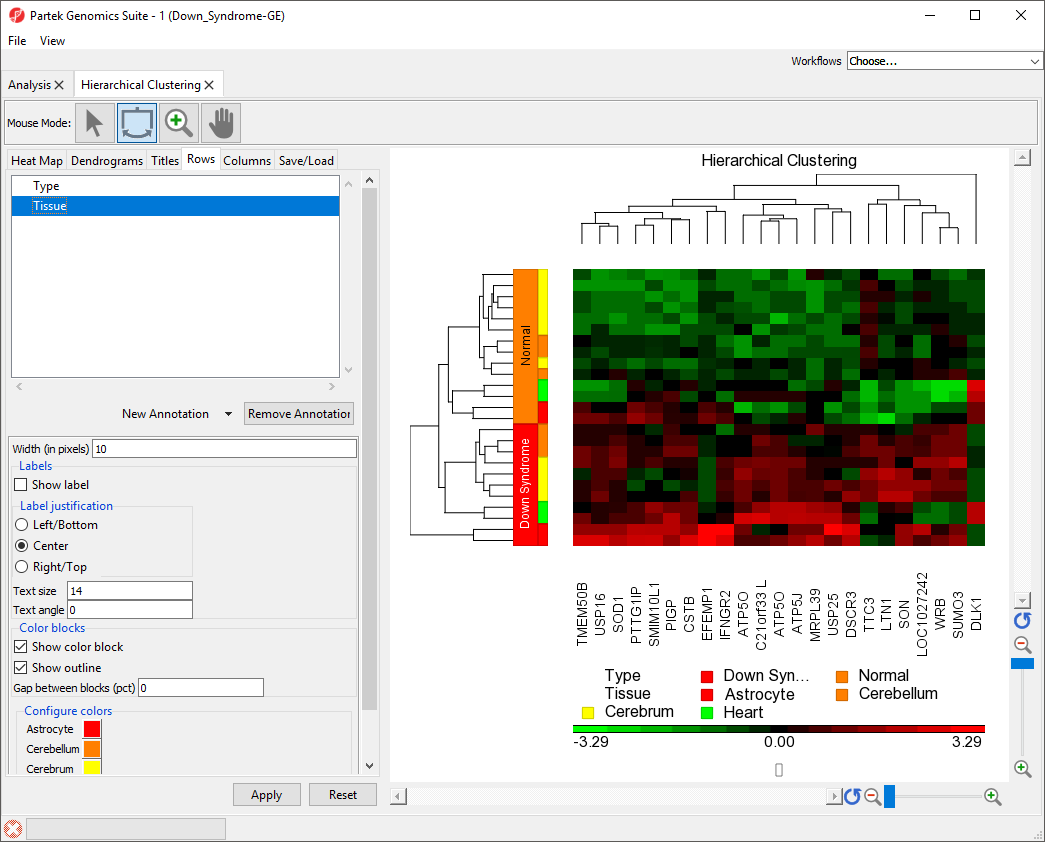
Adding a Sample Attribute to the Heat Map
- Select the Rows tab
- Select Tissue from the New Annotation drop-down menu
- Select Apply
Color blocks indicating the tissue of each sample have been added to the row labels and sample legend (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
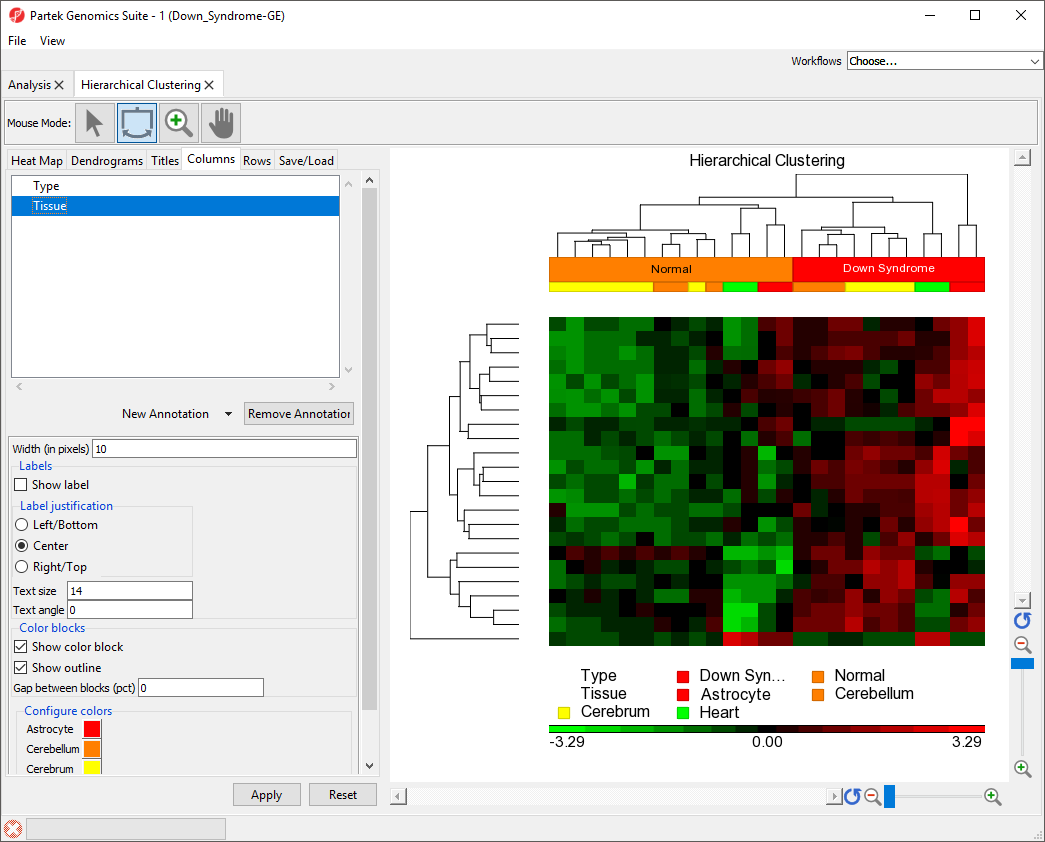
Changing the Orientation of the Rows and Columns
By default, Partek Genomics Suite displays samples on rows and features on columns. We can transpose the heat map using the Heat Map tab in the plot properties panel.
- Select the Heat Map tab
- Select Transpose rows and columns in the Orientation section
- Select Apply
The plot has been transposed with samples on columns and features on rows. The label for the sample groups is now in the vertical orientation because the settings we applied to Rows has been applied to Columns.
- Select the Columns tab
- Select the Type track
- Set Text angle to 0
- Select Apply
The sample group label for Type is now visible (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Flipping Columns or Rows
Each cluster node has two sub-cluster branches (legs) except for the bottom level in the dendrogram, the order of the two branches (or legs) is arbitrary, so the two sub-clusters position can be flipped within the cluster. This does not change the clustering, only the position of the clusters on the plot.
- Select () from the Mouse Mode icon set to activate Flip Mode
- Clicking on a line (or drawing a bounding box on a line using left mouse button) that represents a sub-cluster branch (or dendrogram leg) will flip the selected leg with the other one leg within the same parent cluster. In this example, clicking on the bottom line will move it to the top of the heat map (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
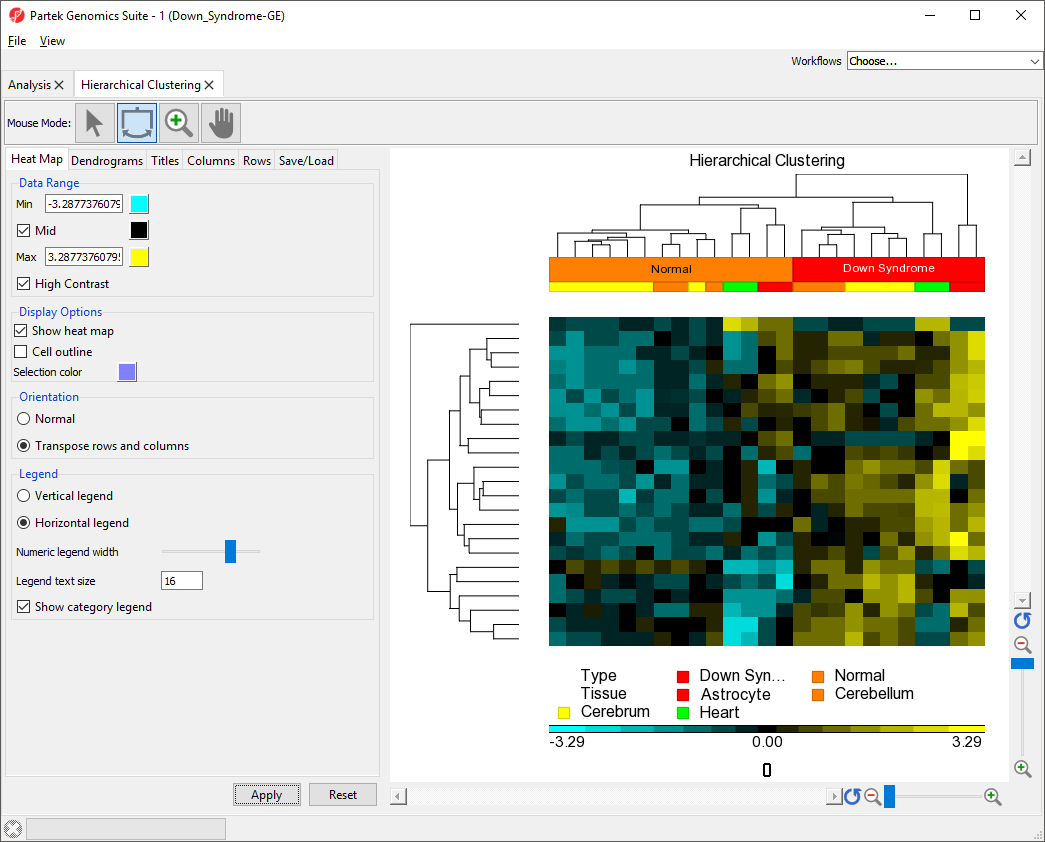
Changing Heat Map Colors
The minimum, maximum, and midpoint colors of the heart map intensity plot can be customized.
- Select the Heat Map tab
- Set Min color to () using the color picker tool
- Set Max color to () using the color picker tool
- Select Apply
The heat map and plot intensity legend now show maximum values in yellow and minimum values in light blue with a black midpoint (Figure 6). The data range can also be customized by changing the values of Min and Max.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
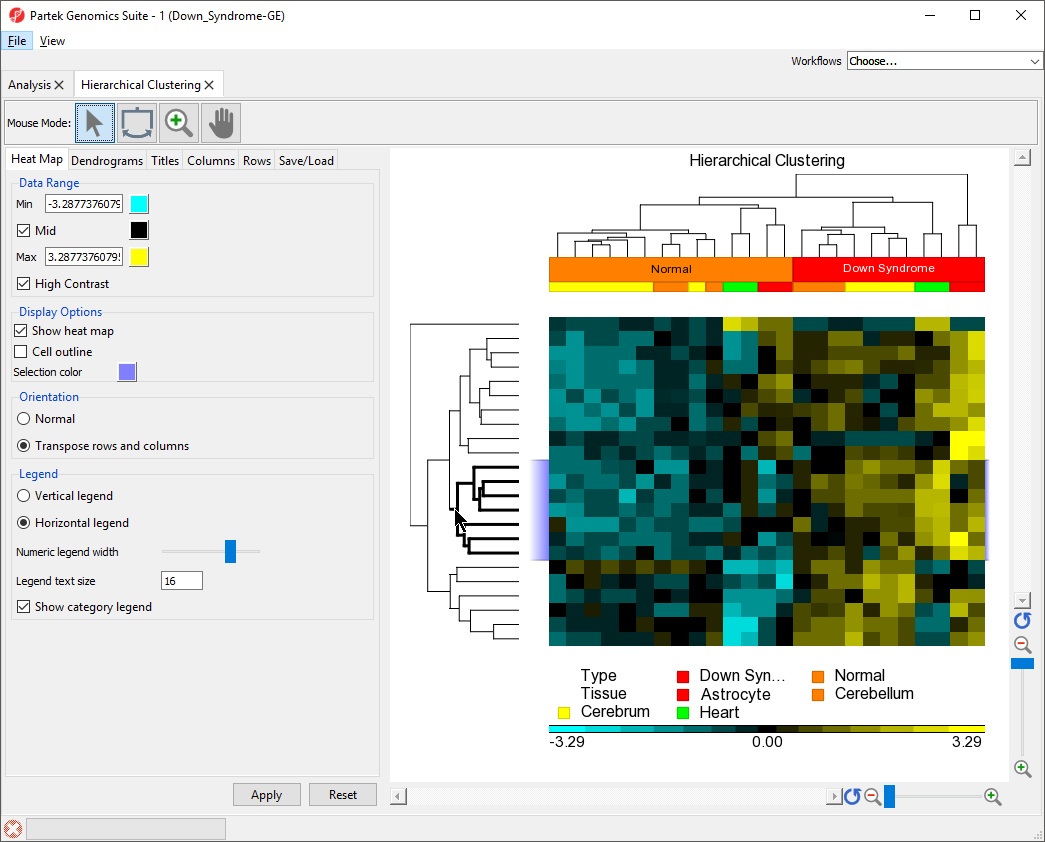
Zooming to Selected Rows/Columns
We can use the hierarchical clustering heat map to examine groups of genes that exhibit similar expression patterns. For example, genes that are up-regulated in Down syndrome samples and down-regulated in normal samples.
- Select () from the Mouse Mode icon set to activate Selection Mode
- Select on the middle cluster of the rows dendrogram as shown (Figure 7) by clicking on the line or drawing a bounding box around the line
The lines within the selected cluster will be bold and the corresponding columns (or rows) on the spreadsheet in the analysis tab will be highlighted.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
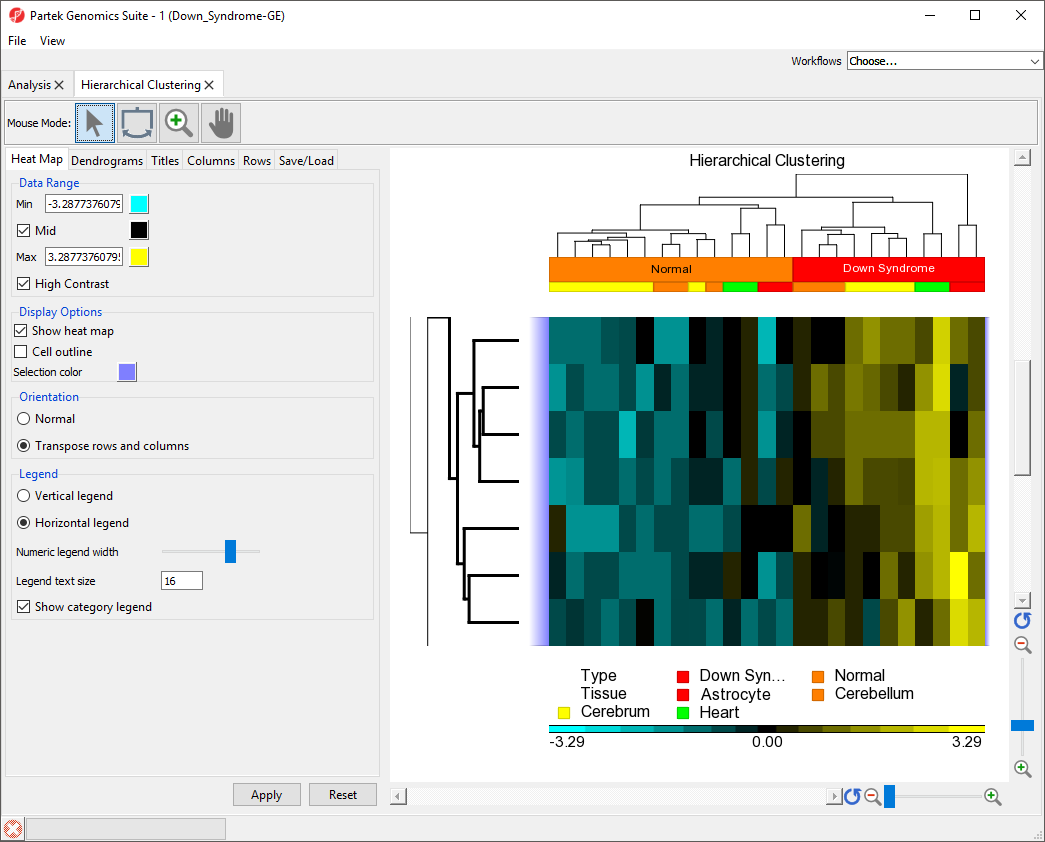
- Right-click anywhere in the viewer
- Select Zoom to Fit Selected Rows
The same steps can be used to zoom into columns or rows. Here, we have zoomed in on rows, but not columns to show the expression levels of the selected genes for all samples (Figure 8).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
To reset zoom select () on the y-axis to show all rows and the x-axis to show all columns.
- Select () on the y-axis to show all rows
- Left click anywhere in the hierarchical clustering plot to deselect the dendrogram
Exporting a List of Genes From a Selected Cluster
Partek Genomics Suite can export a list of genes from any cluster selected, allowing large gene sets to be filtered based on the results of hierarchical clustering.
- Select () from the Mouse Mode icon set to activate Selection Mode
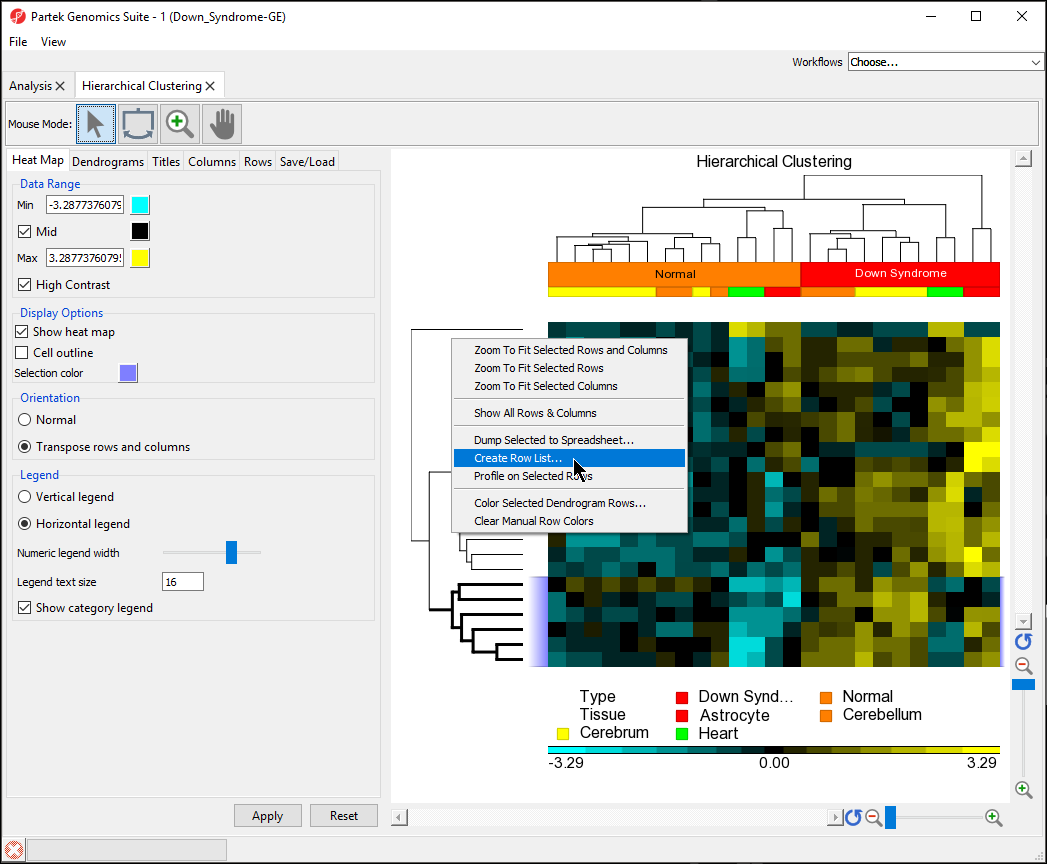
- Select the bottom cluster of the rows dendrogram
- Right-click to open the pop-up menu
- Select Create Row List... (Figure 9)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
- Name the gene set down in normal
- Select OK
- Save the list as down in normal
In the Analysis tab, there is now a spreadsheet row_list (down in normal.txt) containing the 6 genes that were in the selected cluster. The same steps can be used to create a list of samples from the hierarchical clustering by selecting clusters on the sample dendrogram.
Saving Plot Properties
Once you have created a customized plot, you can save the plot properties as a template for future hierarchical clustering analyses.
- Select the Save/Load tab
- Select Save current...
- Name the current plot properties template; we selected Transposed Blue and Yellow
The new template now appears in the Save/Load panel as an option. To load a template, select it in the Load/Save panel and select Load selected. Note that all properties, including Min and Max values and sample groups (based on the column number of the attribute in the source spreadsheet) that may not be appropriate for a different data set, will be applied.
Exporting the Hierarchical Clustering Plot Image
The hierarchical clustering plot can be exported as a publication quality image.
- Select the Hierarchical Clustering tab
- Select File from the main toolbar
- Select Save Image As... from the drop-down menu
- Select a destination and name for the file
- Select PNG or your preferred image type from the pull-down menu
- Select Save
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|