Page History
...
- Select the parent spreadsheet 1 (RNA-seq Tutorial)
- Select Plot Chromosome View in the Visualization section of the RNA-seq Seq workflow panel
Unless you have previously downloaded an annotation file (during another experiment), you will be prompted to select an annotation source.
- Select RefSeq Transcripts - 20142017-0105-0 02
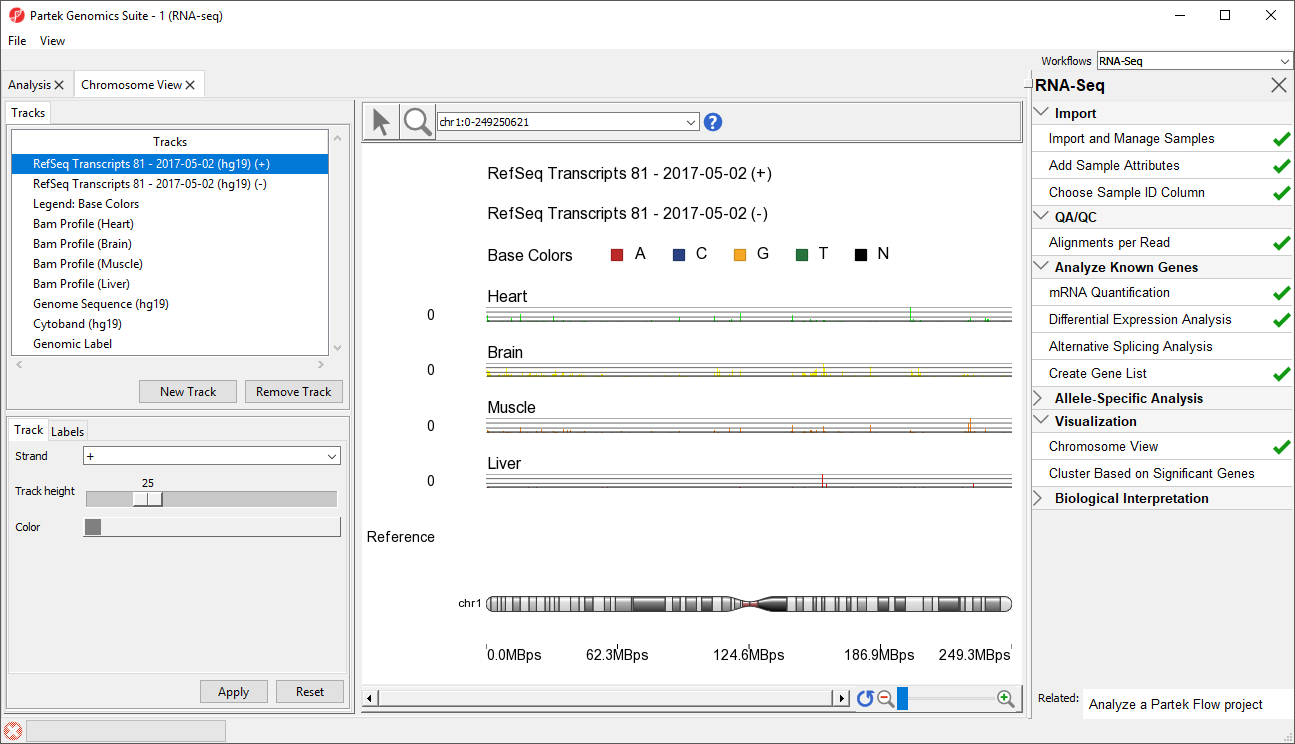
Partek Genomics Suite will download the relevant file and save it to your default library location. The Chromosome View tab will open with chromosome 1 displayed (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
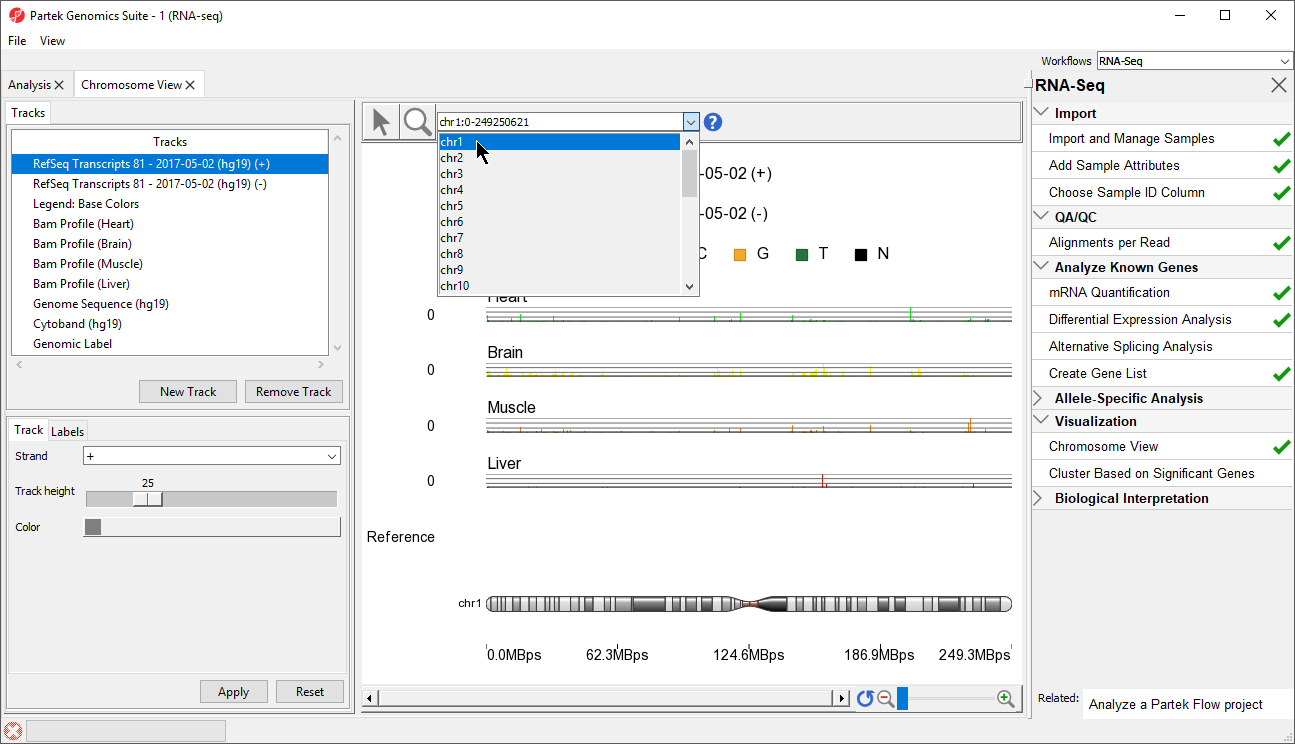
In Chromosome View you can choose other chromosomes from the position field drop-down menu (Figure 2) to change which chromosome is displayed. You may also type a search term (e.g. gene symbol or transcript ID) directly into the position field.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The Tracks panel contains the following tracks:
RefSeq Transcripts (+)
The RefSeq Transcripts (+) track shows all genes encoded on the forward strand of the currently selected chromosome. This experiment uses RefSeq Transcripts, which defines genomic sequences of well-characterized genes, as the reference annotation track. Mouse-over a particular region in this track, and all genes within this region are shown in the information bar. Zoom in on this track to see individual genes, including alternative isoforms.
RefSeq Transcripts (-)
The RefSeq Transcripts (-) track shows all the genes encoded on the reverse strand the currently selected chromosome.
Legend Base Colors
The Legend Base Colors track shows the color for each nucleotide. Colored nucleotide bases become visible in the Bam Profile tracks at higher levels of magnification. By default, the colors are set to red for adenine (A), blue for cytosine (C), yellow for guanine (G), green for thymine (T), and black for base not called (N). The color of the bases can be configured by selecting the Legend Base Colors track and selecting Configure colors in the track configuration panel beneath the Tracks panel on the left-hand side of the Genome Viewer.
Bam Pofile (Heart, Brain, Muscle, Liver)
The Bam Profile tracks show all the reads that mapped to the currently selected chromosome from the four tissue samples. The y-axis numbers on the left side of the tracks indicate the raw read counts. The aligned reads are shown in the Genome Viewer in each track with a different color for each Bam Profile track.
Genome Sequence, Cytoband, and Genomic Label
The Genome Sequence, Cytoband and Genomic Label tracks are shown at the bottom of the panel. These labels are helpful for navigating the chromosome.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|