Page History
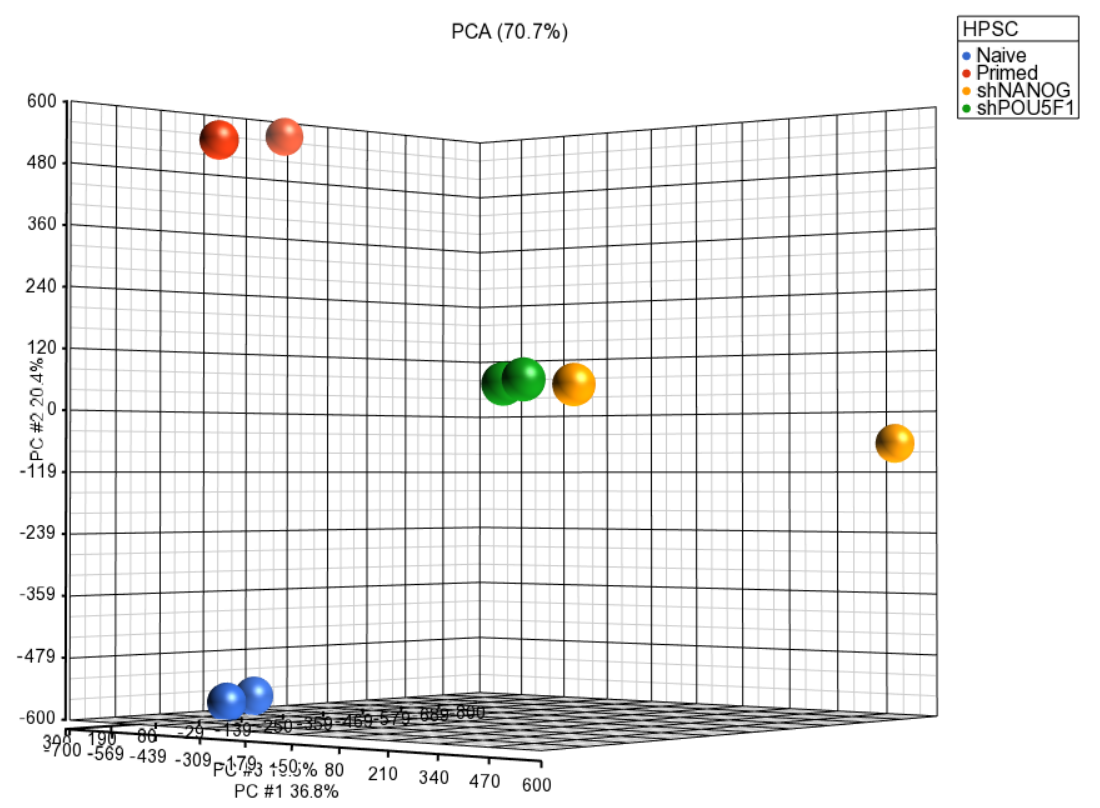
Principal component analysis (PCA) can be invoked on the methylation data to reveal clustering of the samples, but also as a quality control procedure (detection of outliers could point to possible low quality or mislabeled samples). To obtain the PCA plot, switch to the Scatter Plot tab, push Recompute ( ) and from the Color by drop down list select HPSC. Use the Rotate Mode ( )to explore the plot from different angles, as shown seen in Figure 1. Each dot of the plot is a single sample and represents the average methylation status across all CpG loci. The result is shown in the demonstrating clear separation of naive and primed HPSC from the cells transduced with short hairpin (sh) RNA lentiviruses (shNANOG and shPOU5F1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
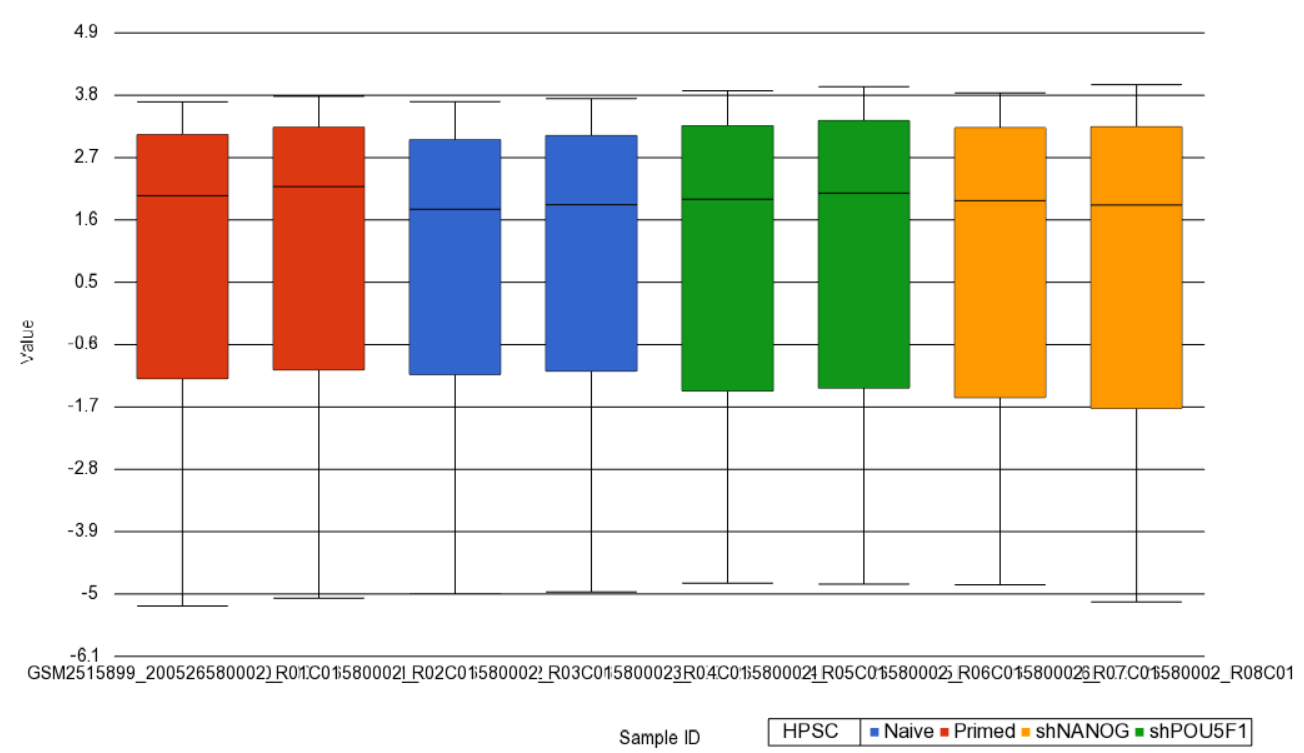
Next, distribution of M-values across the samples can also be inspected by a box-and-whiskers plot: QA/QC > Plot Sample Box & Whiskers Chart. Each box-and-whisker is a sample and the y-axis shows M-values. Samples in this set seem reasonably uniform and no outliers can be detected (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Section Heading
Section headings should use level 2 heading, while the content of the section should use paragraph (which is the default). You can choose the style in the first dropdown in toolbar.
...