Page History
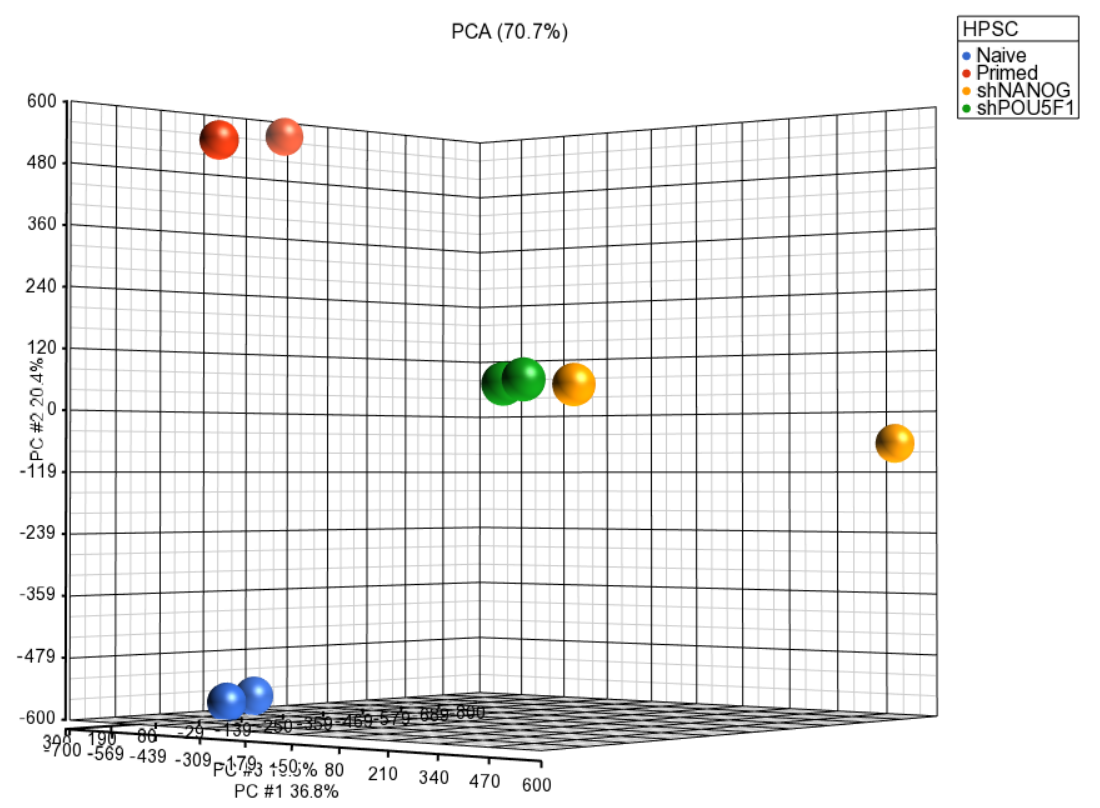
Principal component analysis (PCA) can be invoked on the methylation data to reveal clustering of the samples, but also as a quality control procedure (detection of outliers could point to possible low quality or mislabeled samples). To obtain the PCA plot, switch to the Scatter Plot tab, push Recompute ( ) and from the Color by drop down list select HPSC. Use the Rotate Mode ( )to explore the plot from different angles, as shown in Figure 1. Each dot of the plot is a single sample and represents the average methylation status across all CpG loci. The result is shown in the demonstrating clear separation of naive and primed HPSC from the cells .
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Overview
Content Tools