Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
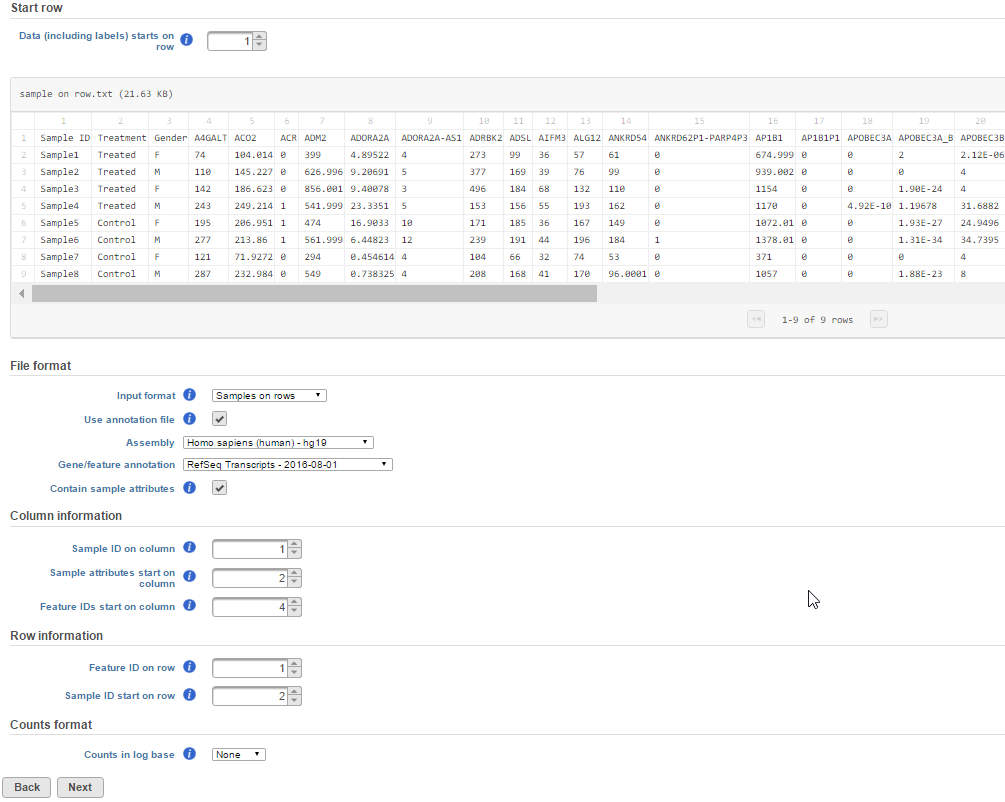
If the read counts are based on a compatible annotation file in Partek Flow, you can specify that annotation file under File formatGene/feature annotation. Select the appropriate genome build and annotation model for your count data. Select the Contain sample attributes checkbox if your data includes additional sample information.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
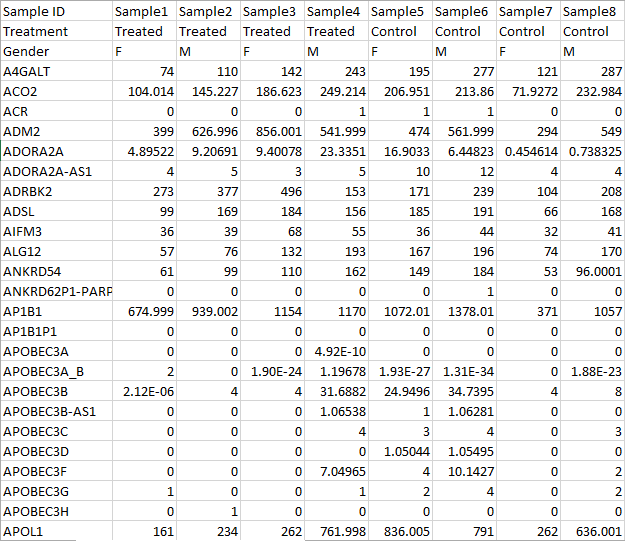
Figure 9 The example above is showing an example text file contains with samples listed on rows, the first column is sample ID, sample attributes are in column 2 and column 3. Gene count starts from column 4 with gene ID in row one. The gene ID . The gene ID is compatible with the hg19 RefSeq hg19 RefSeq Transcripts - 2016-08-01 annotation model. annotation model. Under the Column information and Row information sections, indicate the location of the Sample ID, which in this case is on Column 1. Indicate the sample attribute location by marking where it starts, which in the example is at Column 2. Mark the Feature ID, which in this case are gene IDs and starts at Column 4 .
If the data has been log transformed, specify the base under Counts format.
Project output directory
...