Page History
...
A common task in bulk and single-cell RNA-Seq analysis is to filter the data to include only informative genes. Because there is no gold standard for what makes a gene informative or not, and ideal gene filtering criteria depend on your experimental design and research question, Partek Flow has a wide variety of flexible filtering options.
Filter features task can be invoked from any counts or single cell data node. Noise Reduction and Statistics Based filters take each feature and perform the specified calculation across all of the cells. The filter is applied to the values in the selected data node and the output is a filtered version of the input data node.
In the task dialog, click select the check box filter option to activate one or more of the filter types, type and configure the filter(s), and then click Finish to run.
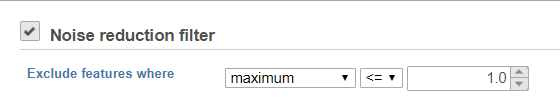
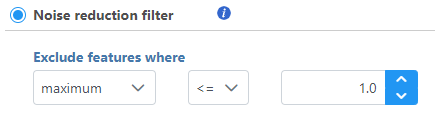
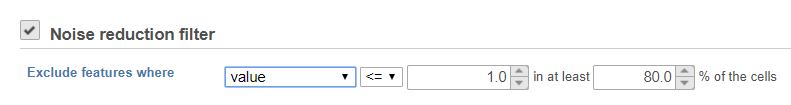
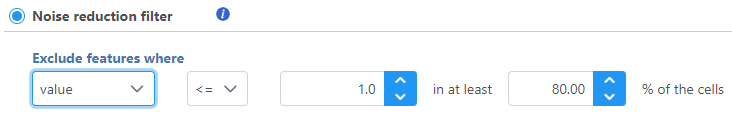
Noise reduction filter
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Descriptive statistics you can choose are:
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
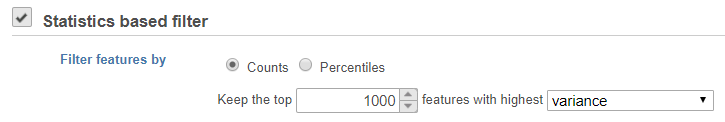
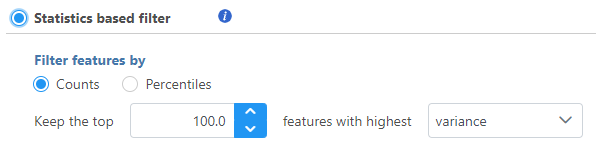
Statistics based filter
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Select Counts to specify a number of top features to include or select Percentiles to specify the top percentile of features to include.
...
- Coefficient of variance
- Geometric mean
- Maximum
- Mean
- Median
- Minimum
- Range
- Standard deviation (std dev)
- Sum
- Variance
- Dispersion
Feature
...
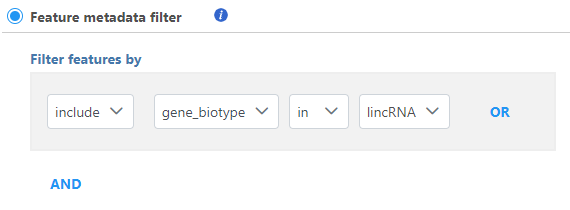
metadata filter
If you have added feature lists in Partek Flow using the List management feature, the Filter list filter will be available (Figure 4the data linked to feature (gene) annotation, different fields in the annotation can be used to filter, e.g. genomic location information, gene biotype information etc (Figure 4)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
You can specify logical operation using different annotation field information.
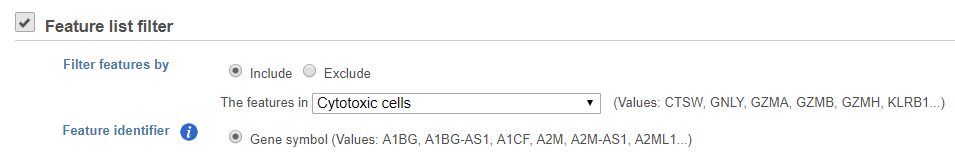
Feature list filter
You can filter features based on a feature lists (Figure 5).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
If you have added feature lists in Partek Flow using the List management feature, the filter using Saved list option will be available. Otherwise, you can specify a Manual list by typing in the Filter criteria box.
If you choose Saved list, the drop-down list will display all the feature lists added in List management; If you choose Manual list, you can manually type in the feature IDs/names in the box, one feature per row.
You can choose to include or exclude features in any list that you have added.
Select the list using the drop-down menu.
Use the Feature identifier option to choose which identifier from your annotation matches the values in the feature list.
...
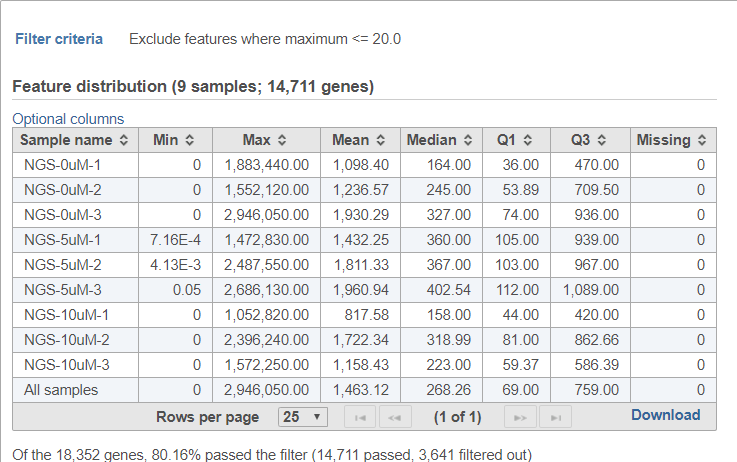
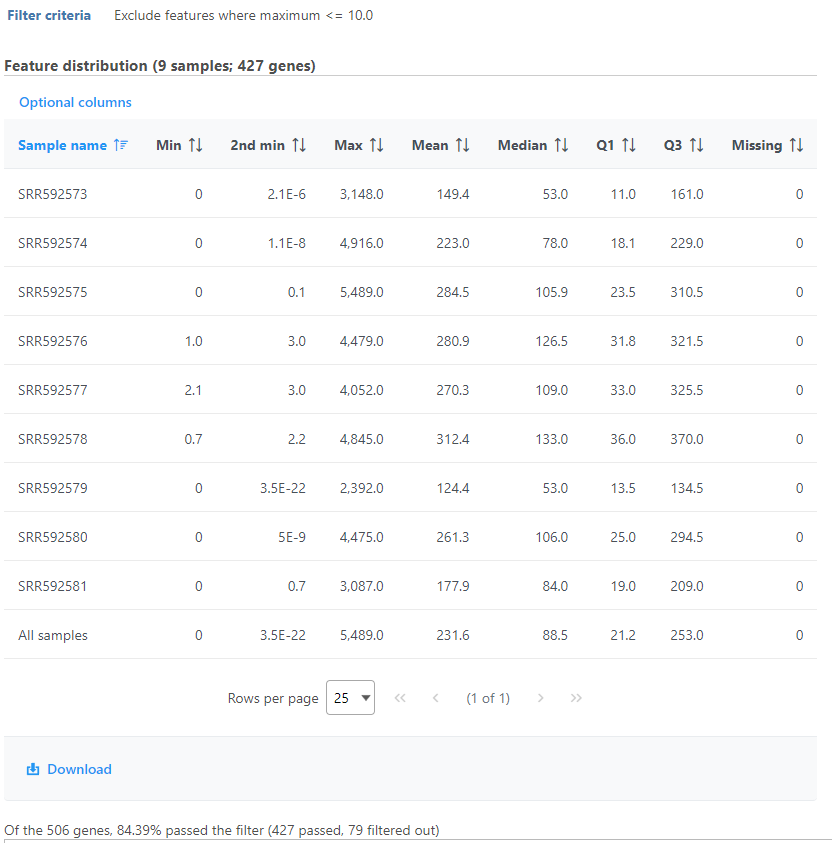
The filter features task report lists the filter criteria, reports distribution statistics for the remaining features, and indicates the number and percentage of features that passed the filter (Figure 6).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
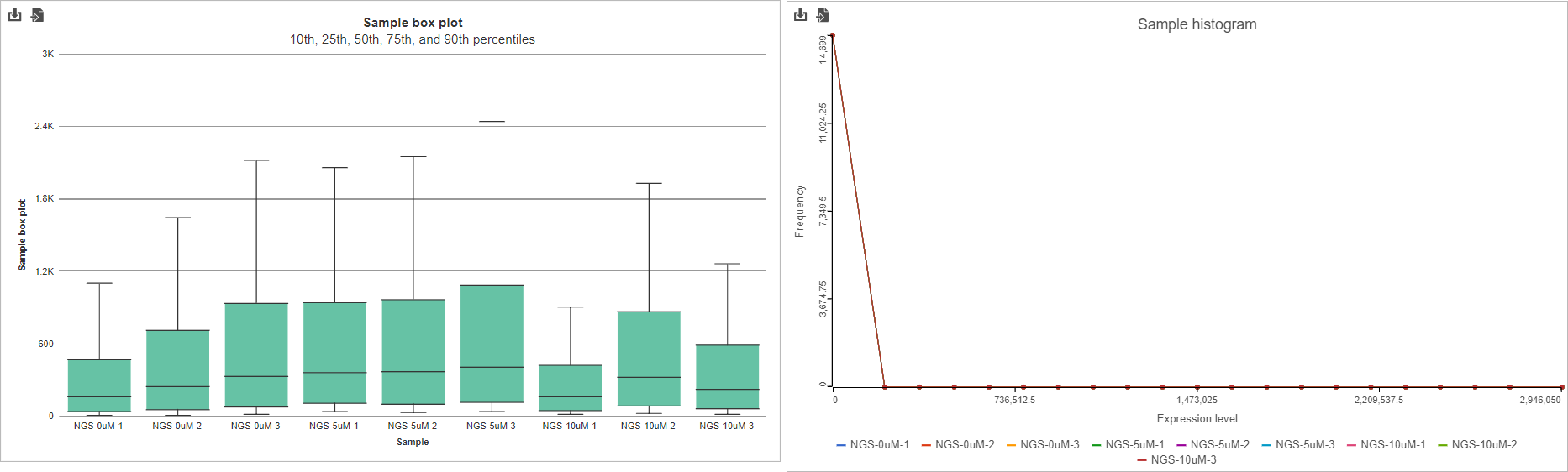
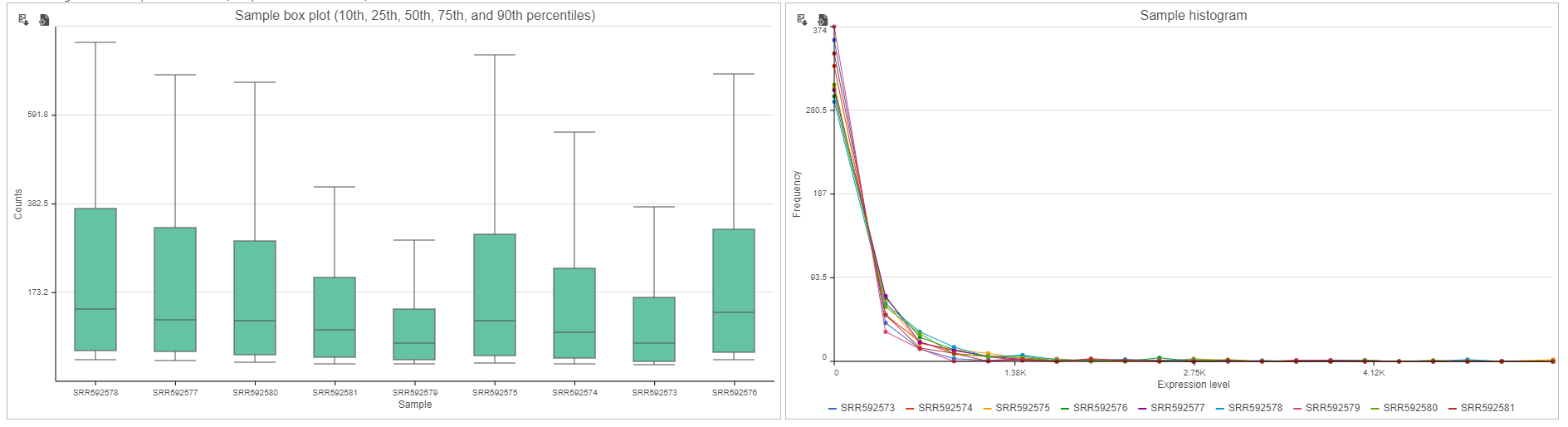
If the input was a count matrix data node, sample box plot and sample histograms are provided to show the distribution of features after filtering. These plots are not available if the input was a single cell counts data node.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
| Additional assistance |
|---|
...