Page History
...
V(D)J recombination occurs in lymphocytes when T and B cells assemble variable (V), diversity (D), and joining (J) gene segments, contributing to the generation of receptors that recognize and respond to perturbations. V(D)J recombination produces clones of unique T cell receptor (TCR) chains or B cell receptor (BCR) chains giving rise to the diverse repertoire of T and B cell populations which are imperative to adaptive immune system function1. The frequency of generated clones can be measured and explored, giving researchers a powerful view into variation, expansion, and diversity within the biological system. You can import filtered Contig Annotation CSV files2 from the 10X 10x Genomics Cell Ranger V(D)J or multi pipeline3. If there is matching gene expression data, it can also be imported and analyzed within the same project. We recommend uploading the filtered feature barcode matrices as either the Hierarchical Data Format (H5 or HDF5)4 or Market Exchange Format (MEX)5.
...
- Create a new project to upload your data. Ensure that you have transferred the filtered contig_annotations.csv file(s)2 for each sample from either the cellranger vdj7 or cellranger multi8 pipeline to the server. If you have matching gene expression data, import the filtered feature barcode matrices as well as in H54 or MEX5 format from the cellranger multi pipeline for each sample.
- Click Import, then select Import single cell V(D)J data
...
- The example below shows the results from the Clonotype Frequency Plot task which is accessed by choosing to perform this task from the Single cell V(D)J node and will automatically open in the data viewer for modification. In this case, the barcode frequency is the number of clonotypes per cell because the barcode usually represents a single cell, so there are two cells that have clonotype5. Clonotype 5 Clonotype5 is made of two compositions (a frequency of four for clonotype5 from the V(D)J node) with a Chain, V gene, D gene, and J gene as seen below the bars and by hovering.
...
- Plotting Clonotype ID frequency from the Cell counts and VDJ node counts nodes highlights the difference between the two nodes. In this example, the top plot is the number of cells per clonotype and the bottom plot is the number of V(D)J clonotypes present. Note that Cell Ranger does not always call the barcode a cell and this can affect frequencies when making comparisons between cell frequency per clonotype and barcode frequency per clonotype. An example of this would be clonotype1 when comparing the figure above and below.
...
- Below in the top chart, CDR3 abundance is plotted by barcode frequency and colored by Clonotype ID in the top Bar chart. In the bottom examplechart, the plot is instead colored by Chain and other modifications, such as axis ticks and the number of groups per page. Note that the predicted CDR3 amino acid sequence is plotted here, but the . The predicted CDR3 nucleotide sequence (cdr3_nt), as well as information for other Complementarity-Determining Regions, is also available.
...
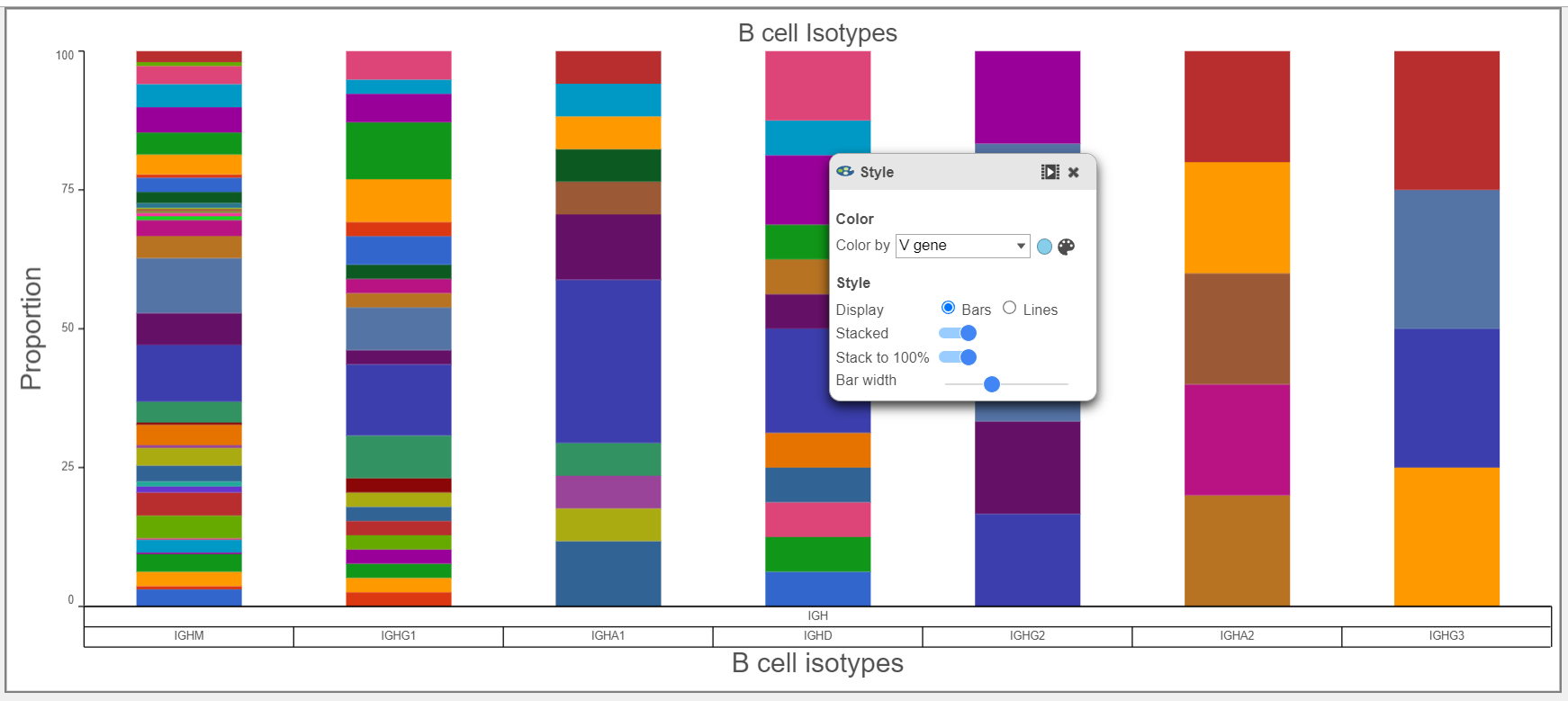
- Gene usage plots for the V, D, and, J genes can be plotted in many ways, as seen by in the Bar chart V gene examples below for the V gene. In the top plot, the barcode frequency for V genes is sorted by frequency in descending order and colored by Chain. The transposed plot below shows all of the groups sorted by ascending value and the heavy chain has been excluded.
...