| Table of Contents |
|---|
| maxLevel | 2 |
|---|
| minLevel | 2 |
|---|
| exclude | Additional Assistance |
|---|
|
Introduction
This tutorial provides information about Partek model selection tool, how to use this function and some common mistakes which we should avoid to do. The dataset used in the tutorial is a simulated human microarray intensity values in log space. The data is not used for diagnostic procedure, only to show how to use the function.
| Table of Contents |
|---|
| maxLevel | 2 |
|---|
| minLevel | 2 |
|---|
| exclude | Additional Assistance |
|---|
|
Select a
...
Classification Model
Download the zip file from the link below here. The download contains the following files:
- Training set data–28 set data: 28 samples (11 disease samples and 15 normal samples) on 9953 genes
- Test set data – data: 8 samples on 9953 genes
- configuration of the model builder (.pcms file)
- 36 sample data set – total of training and test samples
- deployed model (.pbb file)
...
- Open Partek Genomics Suite, choose File>Open File > Open... from the main menu to open the Training.fmt.
Select Tools > Predict > Model Selection from the Partek main menu
- In Cross-Validation tab, choose to Predict onon Type, Positive Outcome is Disease, Selection Criterion is Normalized Correct Rate (Figure 1)
- Choose 1-Level Cross-Validation option, and use Manually specify partition option as 5– use . The idea of 1-level cross validation option is to select the best model to deploy
...
| Numbered figure captions |
|---|
| SubtitleText | Model selection dialog: 1- |
|---|
|
...
| level cross validation configuration | | AnchorName | cross-validation |
|---|
|
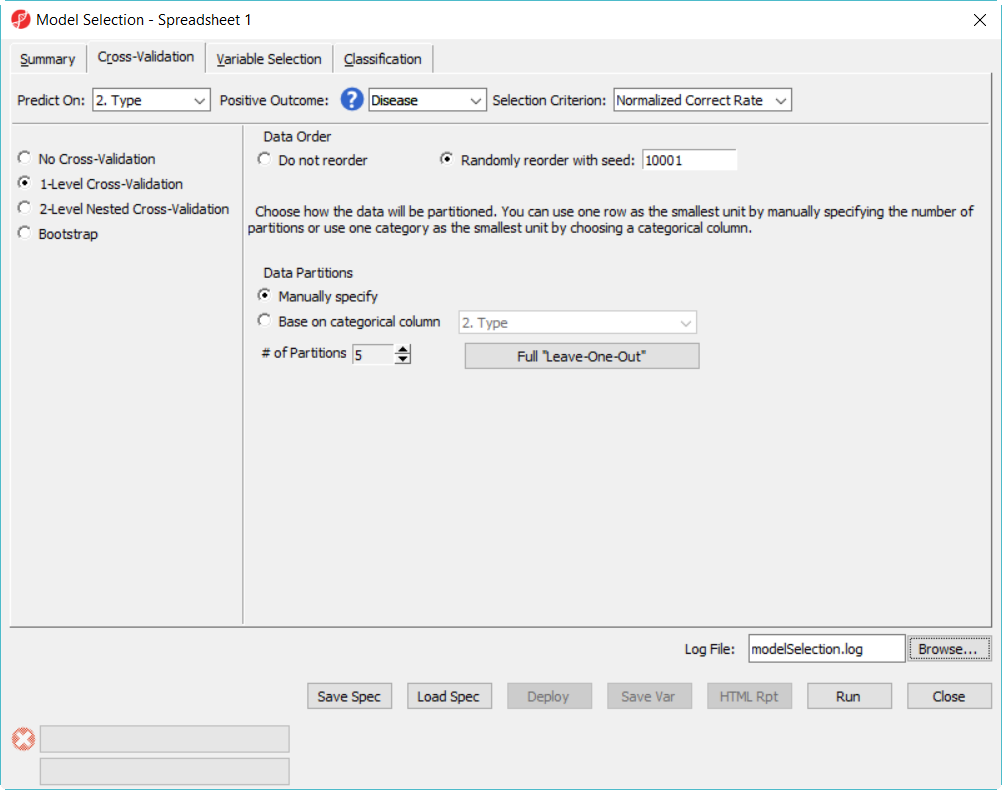 Image Modified Image Modified
|
...
- Choose Variable Selection tab, to use ANOVA to select variables. The number of genes selected are based on the p-value generated from the 1-way ANOVA model which factor is Type. In each iteration of cross validation, we will use the training set to perform ANOVA, take the top N number of genes with the most significant p-values to build the classifier. The Configure button allow you to specify ANOVA model if you want to include multiple factors (Figure 2).
- Since we don't know how many genes should be used to build the model, we will try to use 10, 20, 30, 40, 50 genes – the more options you try, the longer time it takes to run. In the How many groups of variables do you want to try, select Multiple groups with size sizes from 10 to 50 step 10
| Numbered figure captions |
|---|
| SubtitleText | Model selection dialog |
|---|
|
...
| : Variable selection configuration | | AnchorName | varsel |
|---|
|
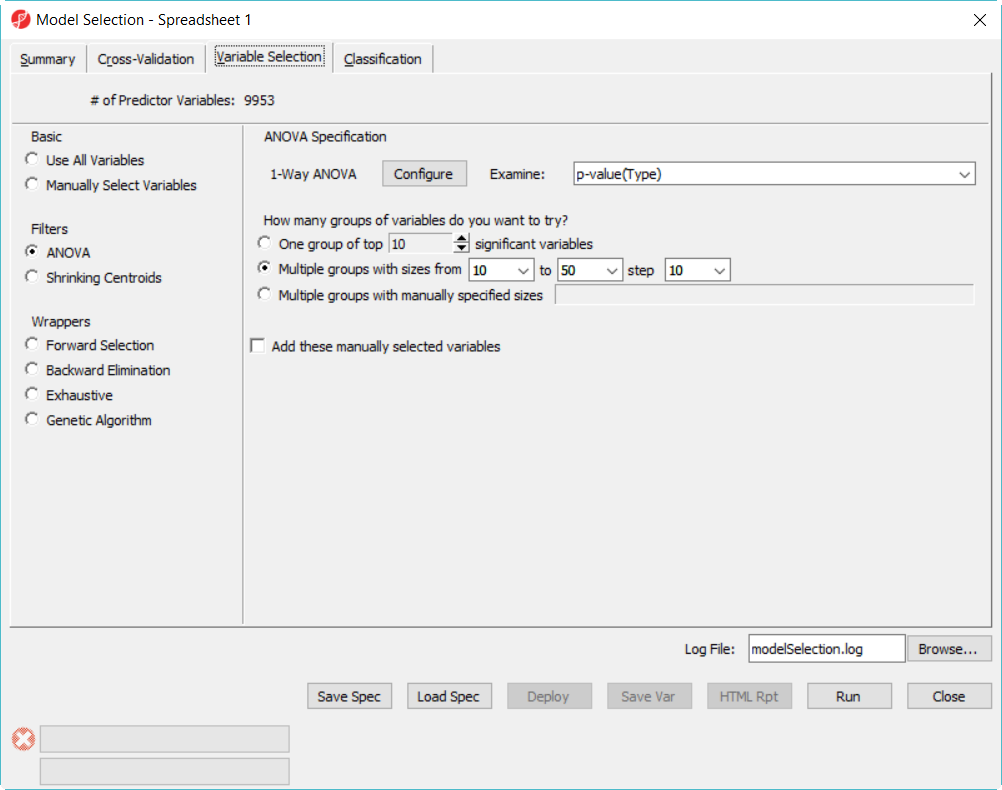 Image Modified Image Modified
|
- Click on Classification tab, select K-Nearest Neighbor, choose 1 and 3 neighbors using default Euclidean distance measure (Figure 3)
...
...
| Numbered figure captions |
|---|
| SubtitleText | Model selection dialog: K- |
|---|
|
...
| nearest neighbor configuration | | AnchorName | knn |
|---|
|
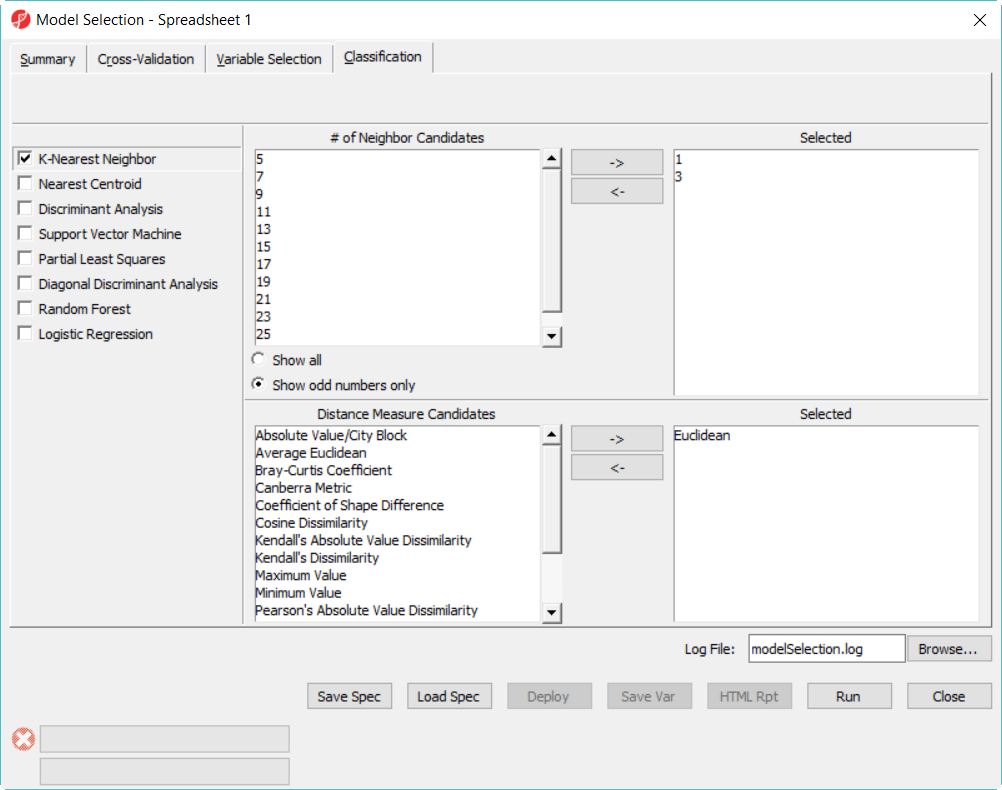 Image Modified Image Modified
|
- Select Discriminant Analysis option, use the default setting which has the Linear with equal prior probabilities option checked
- Click on Summary tab, we have configured 15 models to choose from (Figure 3)
...
...
| Numbered figure captions |
|---|
| SubtitleText | Model selection dialog |
|---|
|
...
| : Summary page | | AnchorName | summary |
|---|
|
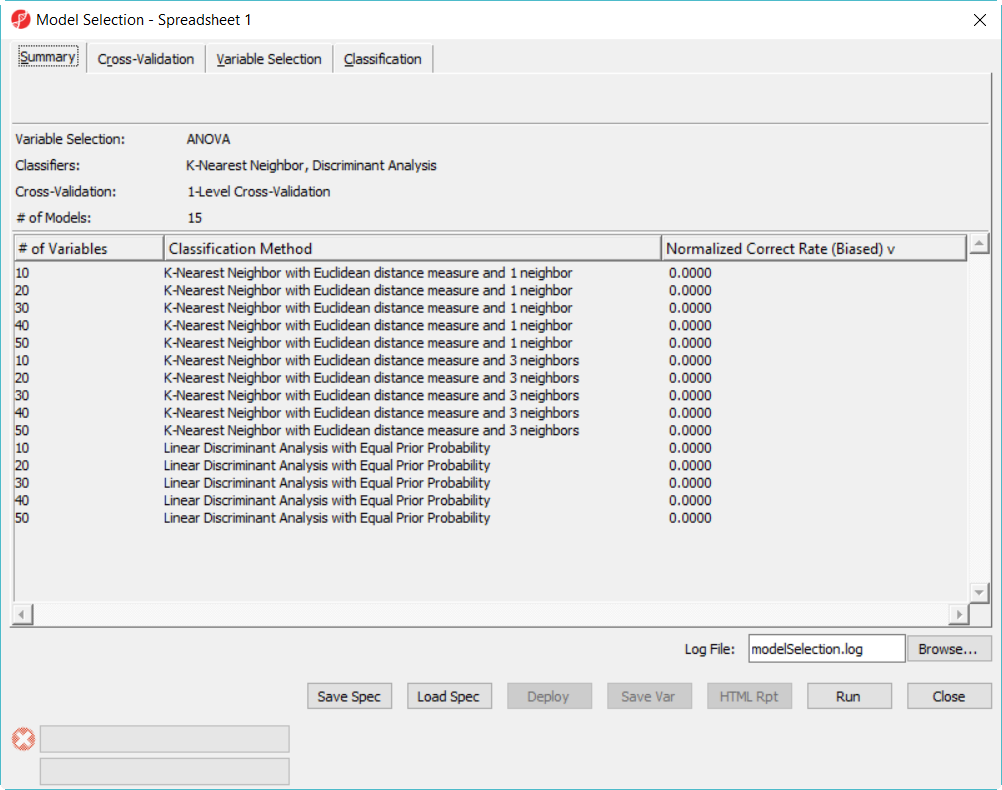 Image Modified Image Modified
|
The more models configured, the long time it takes to run, in this example, in order to save time, we only specified 15 models and choose 5-fold cross-validation. You can also click on Load Spec button to load the above configuration from file tutorial.pcms
When you click on Run, a dialog as (the one in Figure 4) will display, will be displayed, notifying you that some classifiers, like discriminant analysis, are not recommended to perform on dataset with more number of variables than that of samples.
...
| A notification that discriminant analysis model is not recommended |
|
...
...
| with more variables than samples | | AnchorName | curse-of-dimensionality |
|---|
|
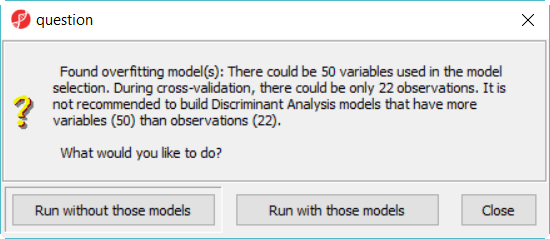 Image Modified Image Modified
|
...
- Click Run without those models button to dismiss the dialog, there will be 12 leaving12 models in this model space
Since we are doing 5-fold cross validation, there will be 6 samples held out as test set in each iteration, and the models are built on the rest remaining 22 samples training set. After it is done, all the 12 models have been tested on the 28 samples, and the correct rate will reported, they are displayed in the summary page in descending order of the normalized correct biased  correct rate, the top one is the best model among the 12 models (Figure 5).
correct rate, the top one is the best model among the 12 models (Figure 5).
...
| One-level cross-validation result: 20 variables 3 nearest neighbor with Euclidean distance measure is the best model among the 12 models on the 28 sample data | | AnchorName | best-model |
|---|
|
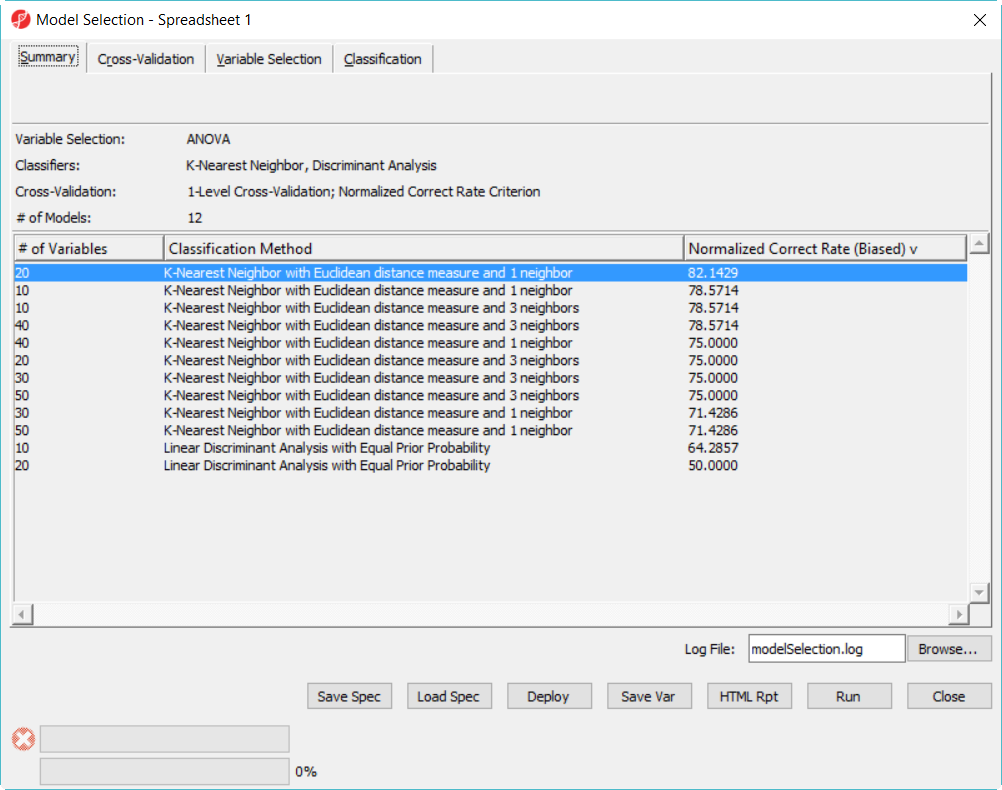 Image Modified Image Modified
|
- Click on Deploy button to deploy leaunch the model using the whole dataset, but first save the file as 20var-1NN-Euclidean.ppb. It will run ANOVA on the 28 samples to generate the top 20 genes and build a model using 3 K-Nearest neighbor based on Euclidean distance measure.
- Since the deployed model was from the whole 28 samples, in order to know the correct rate, we need a test set to run the model on.
...
Deploying a
...
Model
To get unbiased correct rate, the test set sample must be independent from the training set. Now we are going to load another dataset, it has 8 samples with logged intensity values on the set of genes as that of the training data set. To use a complete independent test set to get correct rate is called hold-out validation.
- Choose File>OpenFile > Open... to open to to browse and open testSet.fmt
- Choose Tools>Predict>Run ools > Predict > Run Deployed Model... from the menu
- Select 20var-3NN-Euclidean.ppb to open, click on Test button to run, the correct rate Correct rate (= accuracy) is reported on the top of the dialog (Figure 6)
...
...
...
| Report on deploying a model on a test data set |
|
...
|
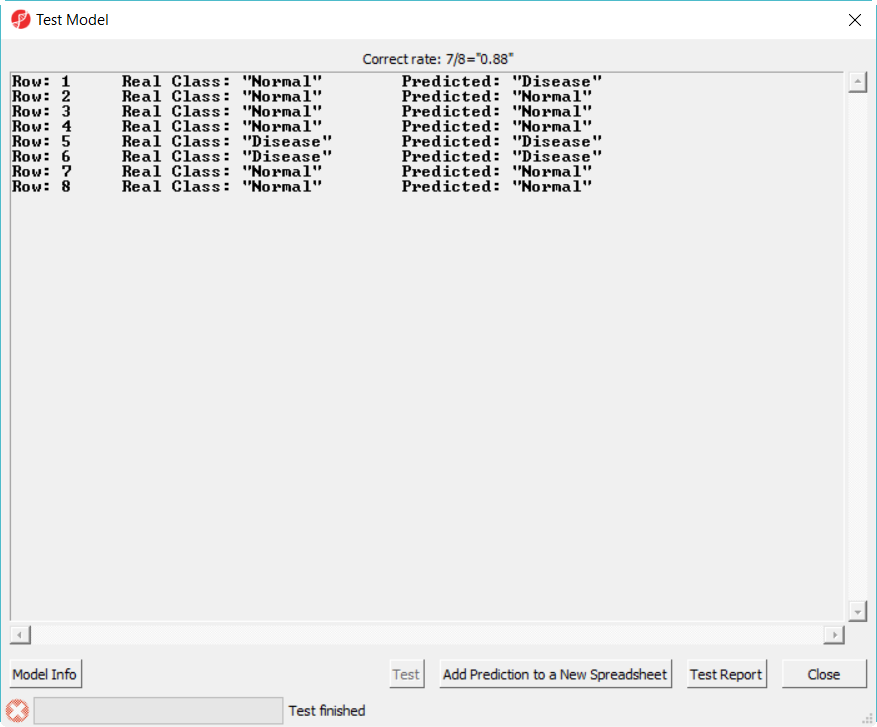 Image Modified Image Modified
|
- Click Add Prediction to New Spreadsheet to generate new spreadsheet with a predicted class name in the first column, the samples (rows) whose predicted and real true class name are different are highlighted (Figure 7)
...
...
| Numbered figure captions |
|---|
| SubtitleText | Test deployed model on test set report on spreadsheet |
|---|
| AnchorName | test_spreadsheet |
|---|
|
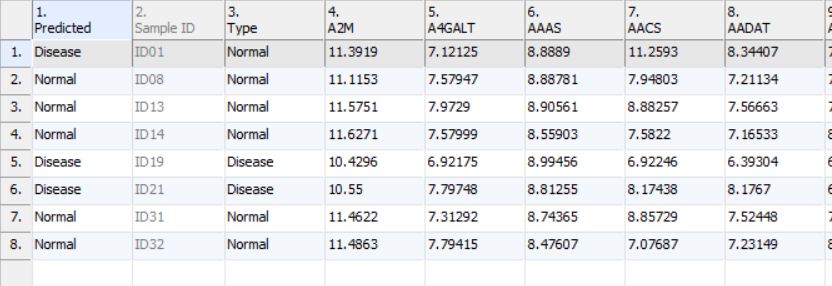 Image Modified Image Modified
|
- Click on Test Report will generate a report in HTML format
- Click Close to dismiss the dialog
Hold-out validation have to split the whole data into two parts -- : training set and test set. Typically However, genomic data (like Microarray microarray or NGS data) typically doesn't have contain a large number of sample sizesamples, os using hold-out method, we have to make the training and test test even smaller, when . When the sample size is small (here the example data is just illustrate the function), the result is not precise. Some people believe that As a rule of a thumb, you should have at least 100 test samples to properly measure the correct rate with a useful precision. The bigger In the other words, the larger the size of training set, the better efficiency of the fitted predicted models are; the bigger size of test set, the better power of validation.
Another method to get unbiased accuracy estimate in Partek Genomics Suite is to do perform a 2-Level Cross validation level cross validation on all the available samples (here: utilize the 36 sample data, samples set), so thta you don't have to split the data. The following steps is to show how to to use all the 36 samples to select the best model and get the accuracy estimate.
- Choose File>OpenFile > Open... to open to browse to and open the file 36samples.fmt
- Choose Tools>Predict>Model ools > Predict > Model Selection... from the menu
- Click on Load Spec to select tutorial.pcms
- Click Run on 1-level cross validation to select the best model using 36 samples
The best model is 30 variables using 1-Nearest Neighbor with Euclidean distance measure (Figure 8).
...
| One-level cross-validation result: 30 variables 1 nearest neighbor with Euclidean distance measure is the best model among the 12 models on the 36 sample data | | AnchorName | best model 36 sample |
|---|
|
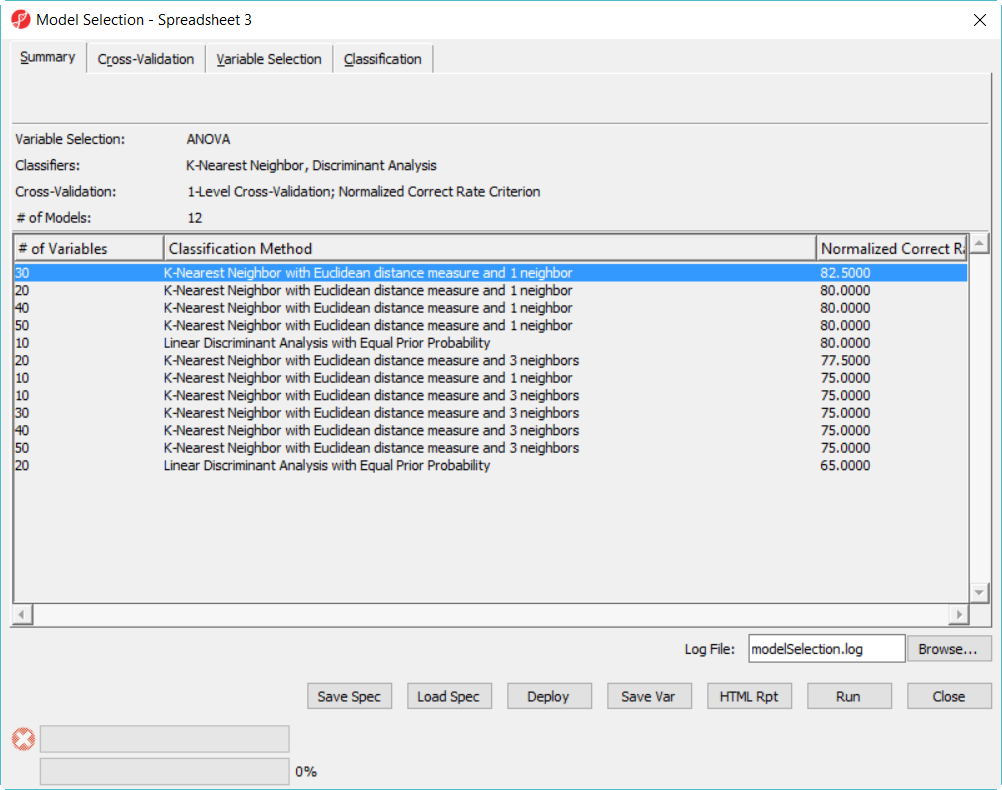 Image Modified Image Modified
|
- Click on the model with best correct rate and deploy the model
...
- Click on Cross-Validation tab, choose 2-Level nested Nested Cross-Validation, specify the number of partition Partition as 5 for both, level level everything else the same and click Run (Figure 9)
...
...
...
| Two-level cross-validation configuration setup | | AnchorName | 2 level cv |
|---|
|
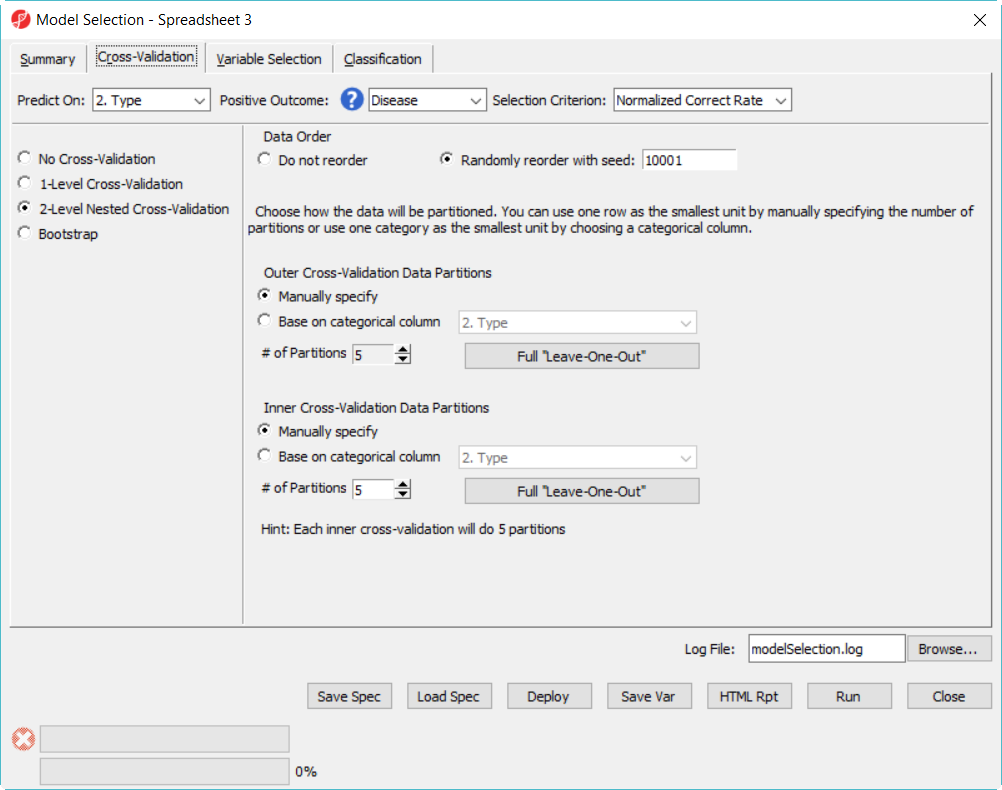 Image Modified Image Modified
|
...
Cross validation
Common mistakes
After it is done, you will get a report like the one in Figure 10. The highligted number is the unbiased accuracy estimate of the best model in the 12 model space.
| Numbered figure captions |
|---|
| SubtitleText | Two level cross-validation report. The highlighted model had the highest accuracy |
|---|
| AnchorName | 2level_report |
|---|
|
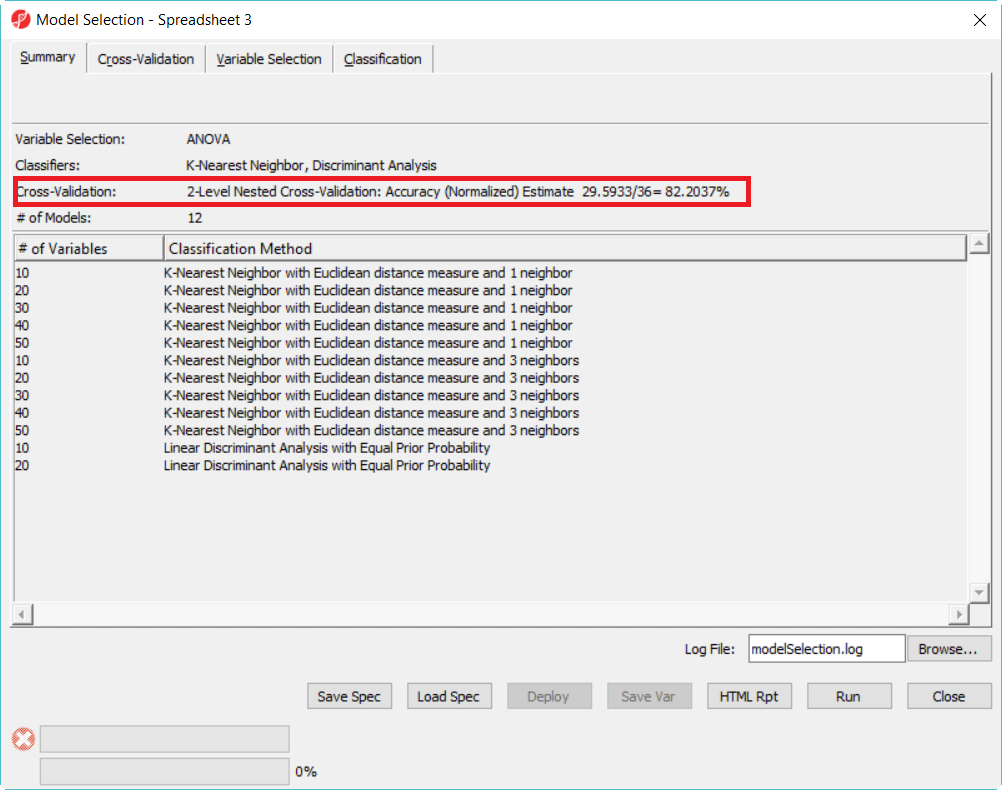 Image Added Image Added
|
Cross-validation
Cross validation is used to esimate the accuracy of the predictive model, it is the solution to overfitting problem. One example of ovefitting is testing the model on the same data set when the model is trained. It is like to give sample questions to students to practice before exam, and the exact same questions are given to the students during exam, so the result is biased.
In cross-validation, the data is partition the data into training set and test set, build a model on training set and validate the model on the test set, so test set is completely independing from model traininig. An example of K-fold cross-validation is to randomly divide the whole data into k equal sized subsets, take one subset as test set, use the remanining k-1 subset to training a model, it will repeat K times, so all the samples are used for both training and test, and each sample is tested once. The following figure is showing 5-fold cross-validation:
| Numbered figure captions |
|---|
| SubtitleText | 5-fold cross-validation |
|---|
| AnchorName | cv |
|---|
|
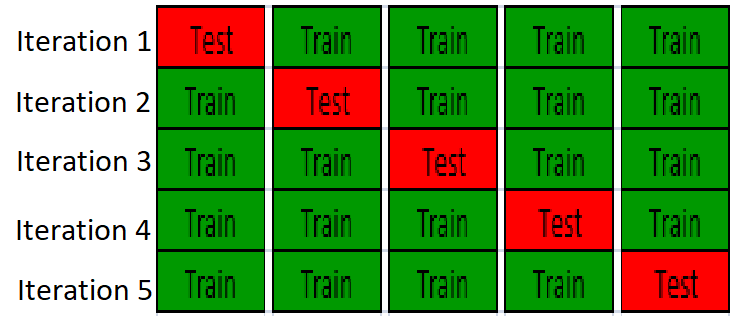 Image Added Image Added
|
Common Mistakes
In Partek model selection tool, the cross-validation is performed first. Each iteration of cross-valiation, the variable selection and classification are performed on the training set, and the test set is completely independent to validate the model. One common mistake is to select variable beforehand, e.g. using perform ANOVA on the whole dataset and use ANOVA's result to select top genes, and perform the cross-valiation to get correct rate. In this case, the test sets in cross validation were used in the variable selection, it is not independend from the training set, so the result will be biased.
Another common mistake is to run 1-level cross-validation with multiple models, and report the correct rate of the best model as the estimate of generalization correct rate, This correct rate is optimistically biased. The reason is that in 1-level cross validation, the test set is used to select the best model, the test set is not independent anymore in terms of estimating correct rate on a unseen dataset. So either use 2-level cross-validation option or use another independ set to get the accuracy estimate, the idea here is to partition the data into 3 sets: training set, validation set and test set. Train the models on the training set, validation set is used to select the best model, and test set is used to generate an unbiased accuracy estimate.
...
correct rate, the top one is the best model among the 12 models (Figure 5).











