Page History
...
Discover de novo motifs
- Select Motif discovery Discovery from the Peak Analysis section of the ChIP-Seq workflow
- Select Discover de novo motifs
- Select OK
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
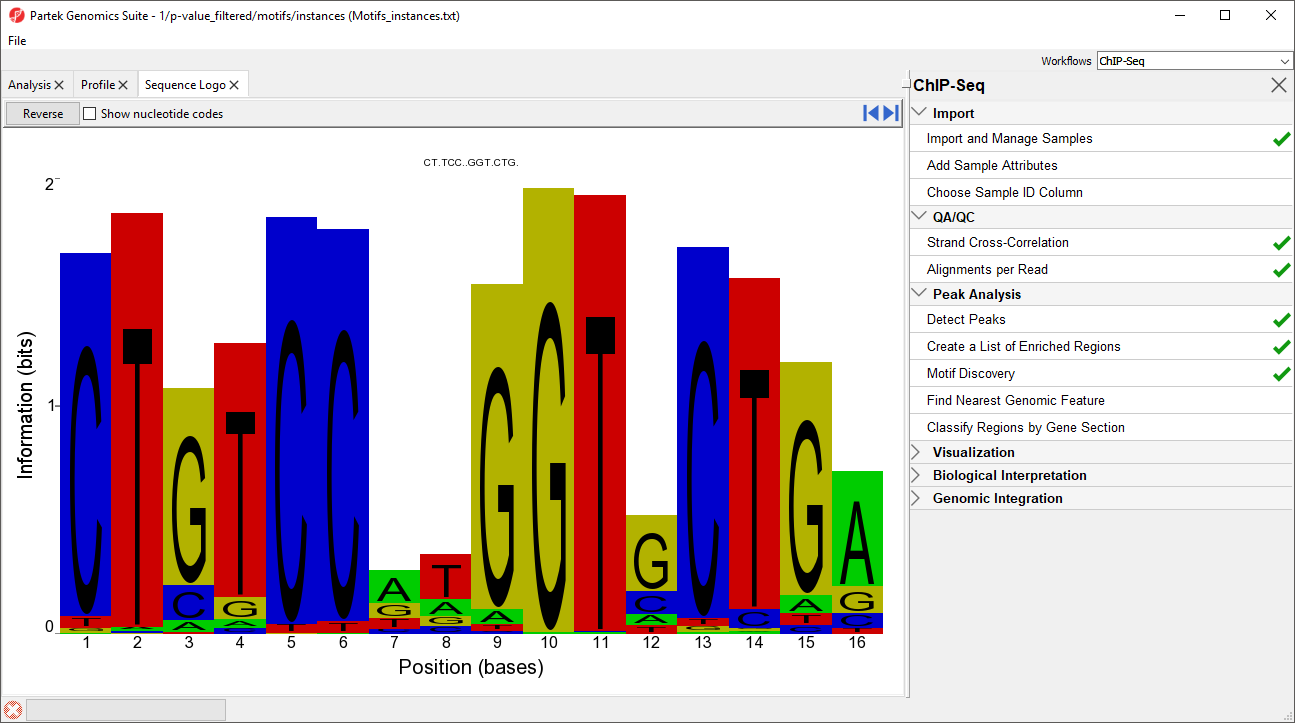
In this case, the motif finder discovered a motif in the NRSF-enriched regions that is 16 base pairs in length. The height of each position is the relative entropy (in bits) and indicates the importance of a base at a particular location in the binding site.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
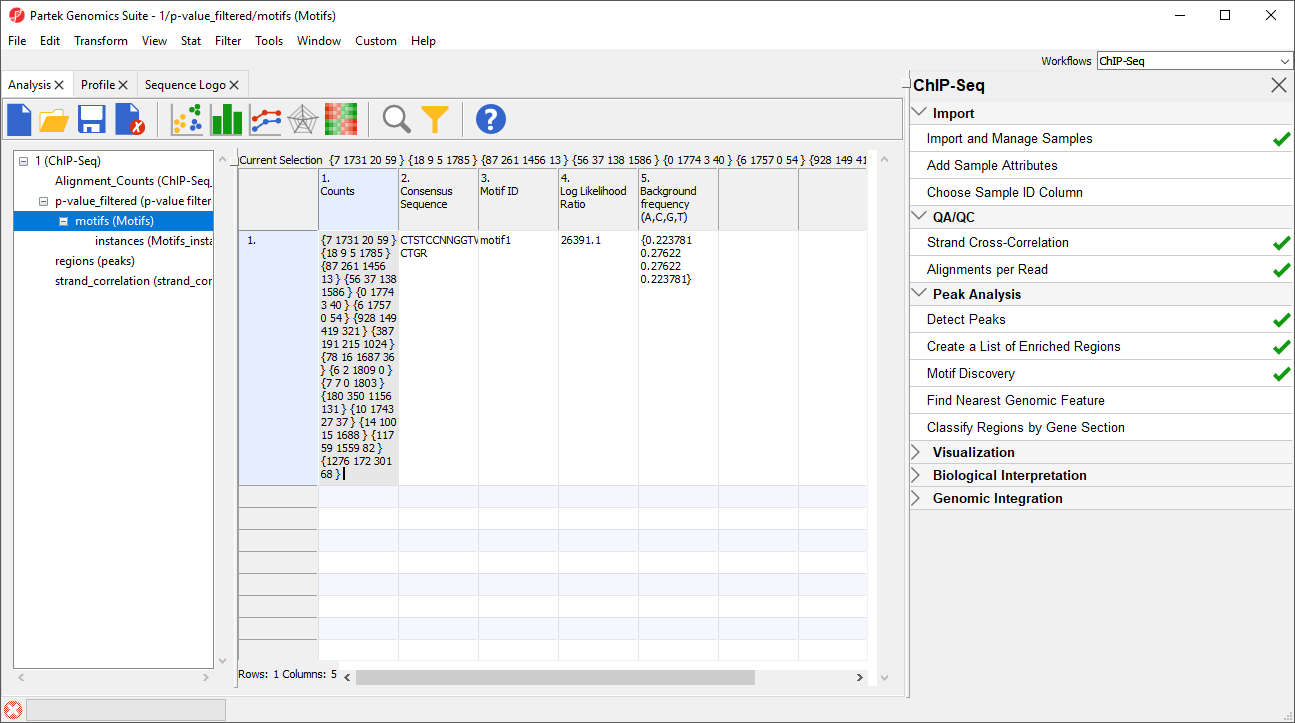
1. Counts gives the summed counts for each base call across all occurrences of the motif in the region list as {A, C, G, T}
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
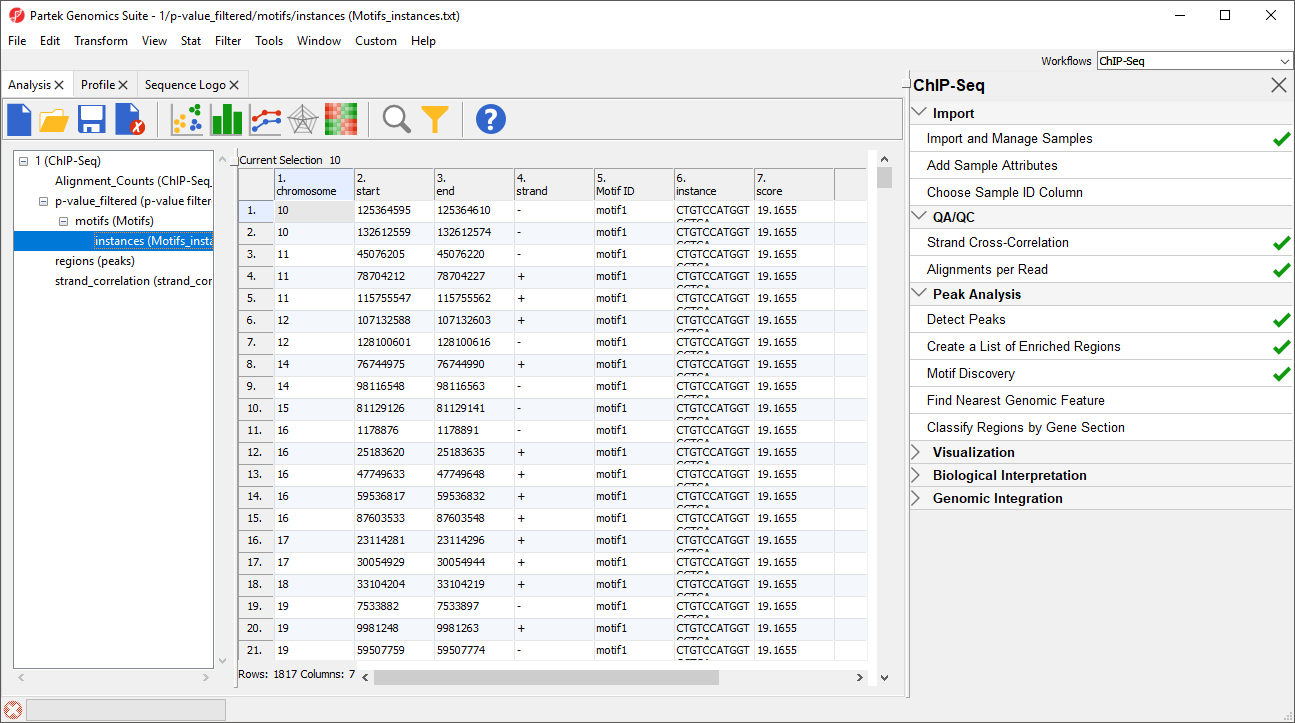
1-4. chromosome, start, stop, strand give the position
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
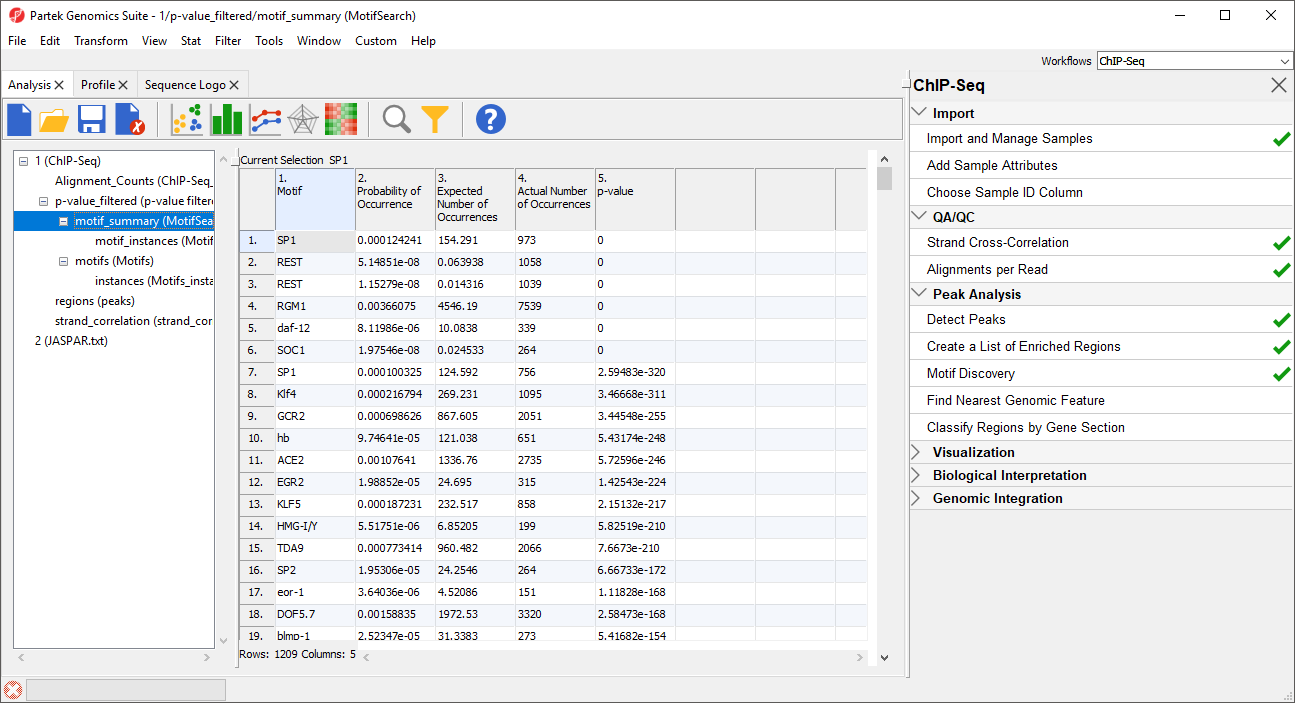
In the MotifSearch spreadsheet, each motif used in the motif search is shown. The columns detail the results of the search for each motif that was found in the reads.
...
Neuwald, A. F., Liu, J.S., & Lawrence, C.E. (1995). Gibbs motif sampling: detection of outer membrane repeats (Vol. 4). Protein Science.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|