Page History
SC transform task performs the normalization method of R package sctransform [1]variance stabilizing normalization proposed in [1]. The task's interface follows that of SCTransform() function in R [2]. SCTransform v2 [3] provides the ability to perform downstream differential expression analyses besides the improvements on running speed and memory consumption. v2 is the default method in Flow.
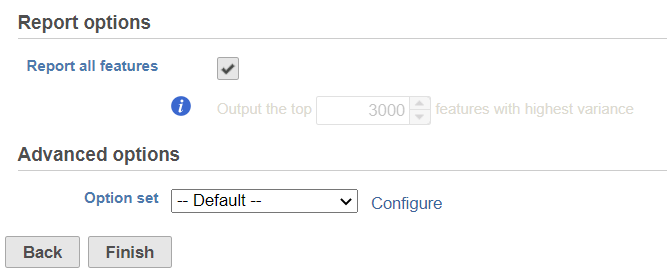
We recommend performing sctransform the normalization on a single cell row raw count data node. Select SCTransfrom SCTransform task in Normalization and scaling section on the pop-up menu to invoke the dialog (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
By default, it will generate report on all the input features. Unchecking the Report all features, user can specify limit the results to a certain number of features with highest variance in the report.
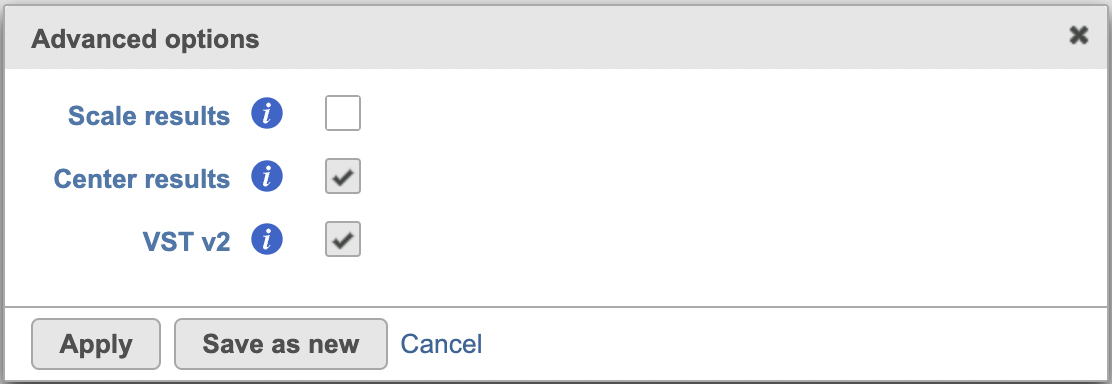
In the Advanced optionoptions, users can the click Configure to change the default settings (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
Scale results: Whether to scale residuals to have unit variance; default is FALSE
...
Center results: When set to Yes, center all the transformed features to have mean as 0
Clip results: If this is set to No, outliers might have big effect and the transformed data can be very large for some features, usually the ones with few non-zero counts. When set to Yes, the range to clip the transformed data is between -sqrt(n/30) and sqrt(n/30), n is the number of cells
Random seed: use the same random seed to reproduce the results.
Data has been log transformed with base: specify the input data is logged or not
The data in the output node is a matrix of standardized residual on all the features in all the observations, the range of the values is roughly between -4 and 4.
References
zero mean expression. Default is TRUE.
VST v2: Default is TRUE. When set to 'v2', it sets method = glmGamPoi_offset, n_cells=2000, and exclude_poisson = TRUE which causes the model to learn theta and intercept only besides excluding poisson genes from learning and regularization; If default is unchecked, it uses the original sctransform model (v1), it will only generate SC scaled data node.
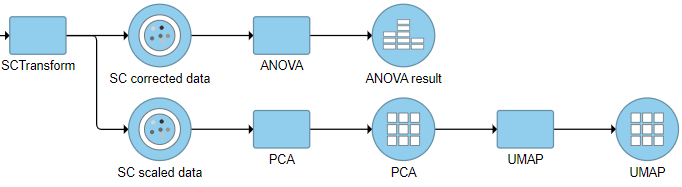
There are two data nodes generated from this task (if VST v2 option is checked as default):
SC scaled data: it is a matrix of normalized values (residuals) that by default has the same size as the input data set. This data node is used to perform downstream exploratory analysis e.g. PCA, Seurat3 integration etc (Figure 3), this data node is not recommend to use for differential analysis.
SC corrected data: is equivalent to the ‘corrected counts’ in data slot generated after PrepSCTFindMarkers task in the SCT assay in Seurat object. It is used for downstream differential expression(DE) analyses (Figure 3).
Note: When perform DE analysis with Hurdle, the 'shrinkage of error term variance' option might need to turn off depending on the dataset. Similarly, the 'Lognormal with shrinkage/voom' option needs to turn off when run DE with GSA.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
References
- Christoph Hafemeister, Rahul Satija. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. https://doi.org/10.1101/576827
- SCTransform() documentation https://www.rdocumentation.org/packages/Seurat/versions/3.1.4/topics/SCTransform
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|