Page History
...
Discover de novo motifs
- Select Motif discovery Discovery from the Peak Analysis section of the ChIP-Seq workflow
- Select Discover de novo motifs
- Select OK
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
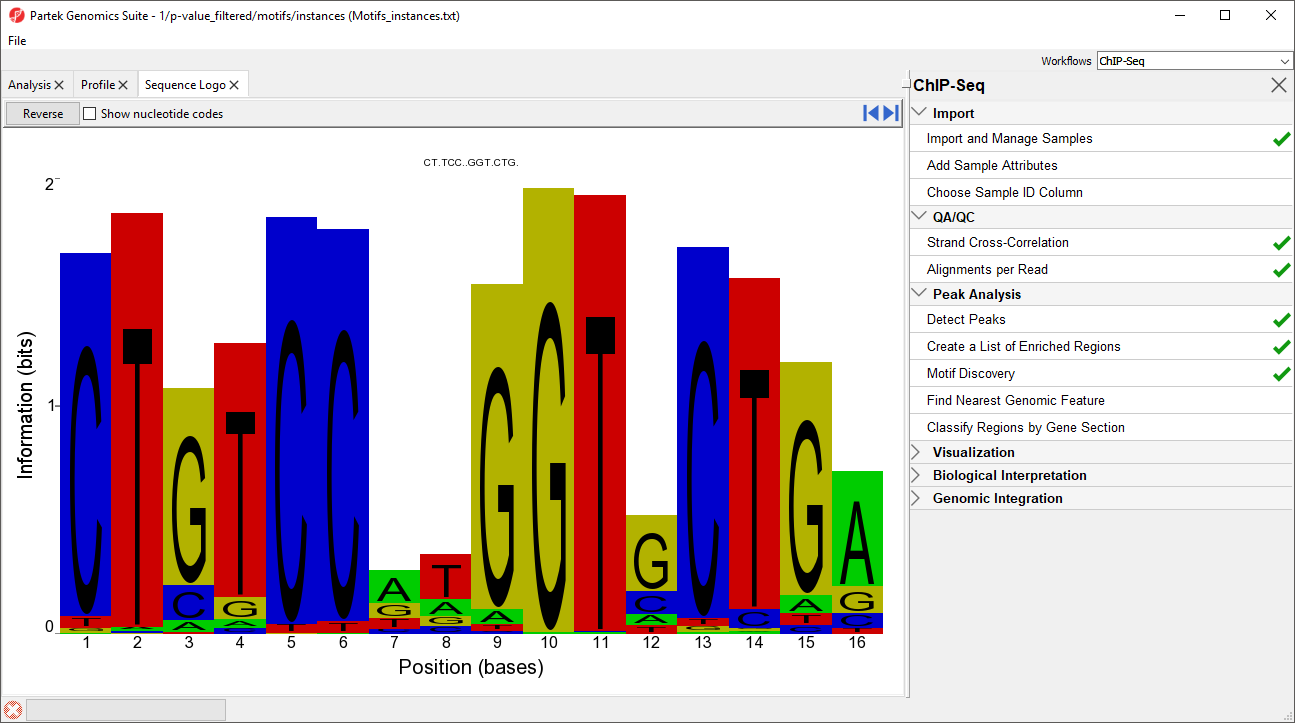
In this case, the motif finder discovered a motif in the NRSF-enriched regions that is 16 base pairs in length. The height of each position is the relative entropy (in bits) and indicates the importance of a base at a particular location in the binding site.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
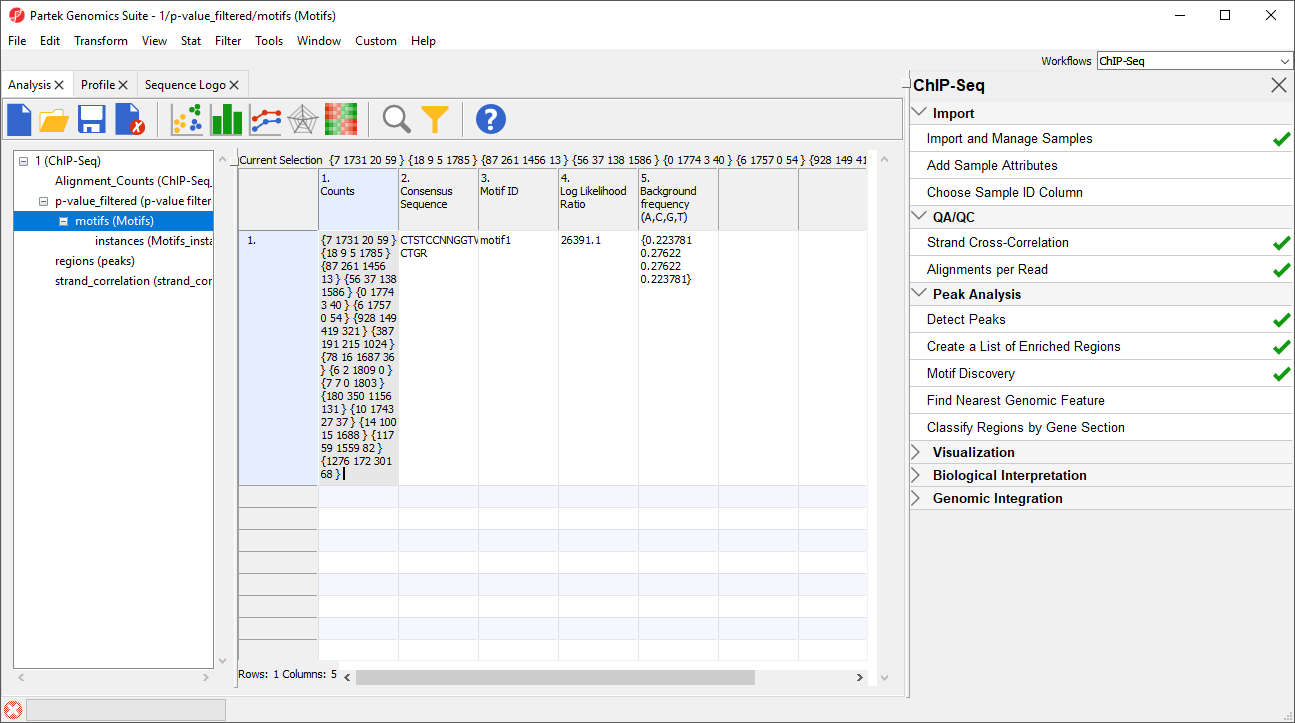
1. Counts gives the summed counts for each base call across all occurrences of the motif in the region list as {A, C, G, T}
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
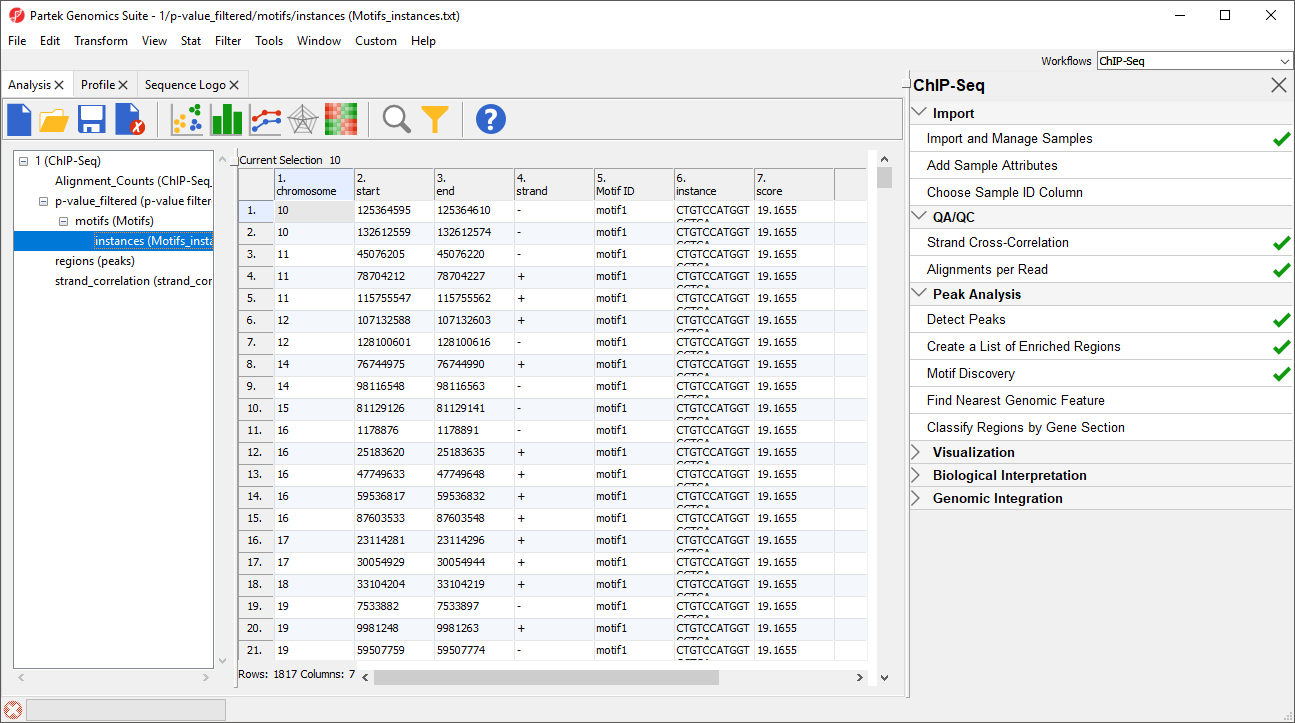
1-4. chromosome, start, stop, strand give the position
...
Two spreadsheets are created, similar to the spreadsheets in the de novo motif discovery, the motif_summary (MotifSearch) spreadsheet (Figure 67) and the motif_instances (MotifSearch.instance) spreadsheet.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
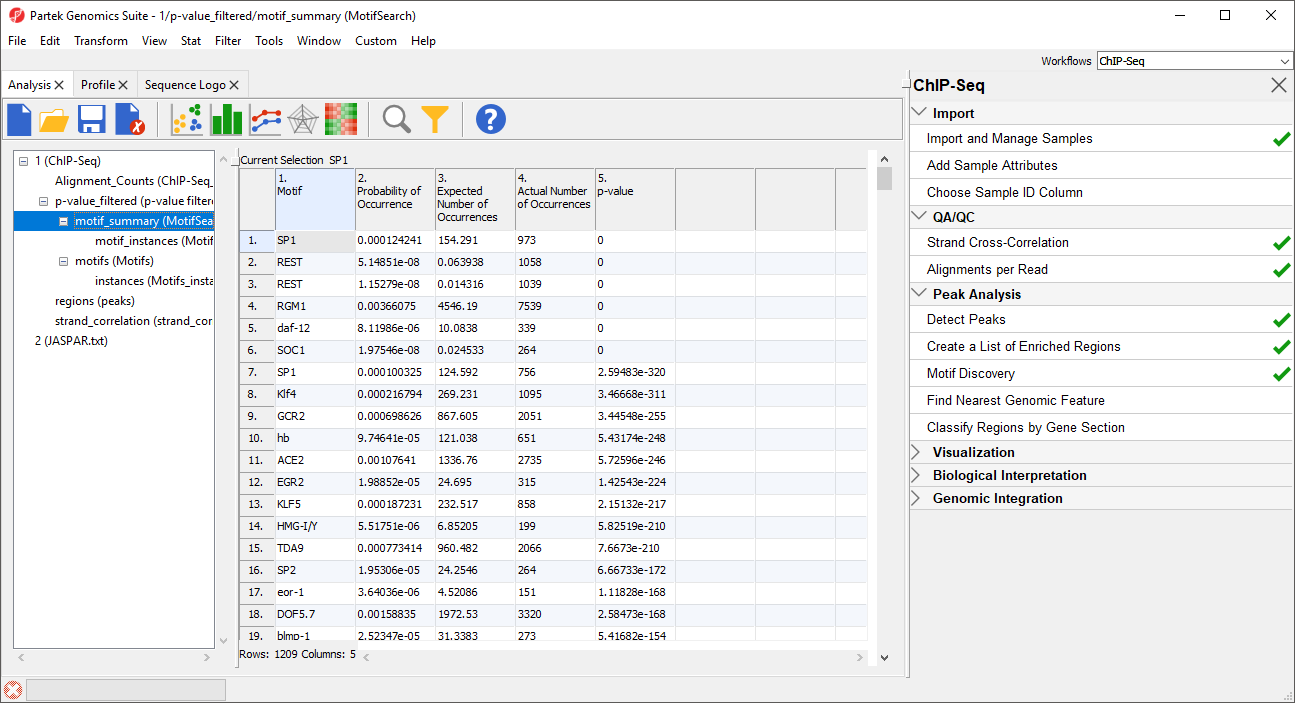
In the MotifSearch spreadsheet, each motif used in the motif search is shown. The columns detail the results of the search for each motif that was found in the reads.
...
As you can see, REST, which is another name for NRSF, is near the top of the list as one of the most significantly over-represented motifs (Figure 67). This motif agrees with the motif found in the de novo motif detection step. Interestingly, other motifs appear a significant number of times in the ChIP-Seq peaks and may represent possible co-factors or regulators.
...
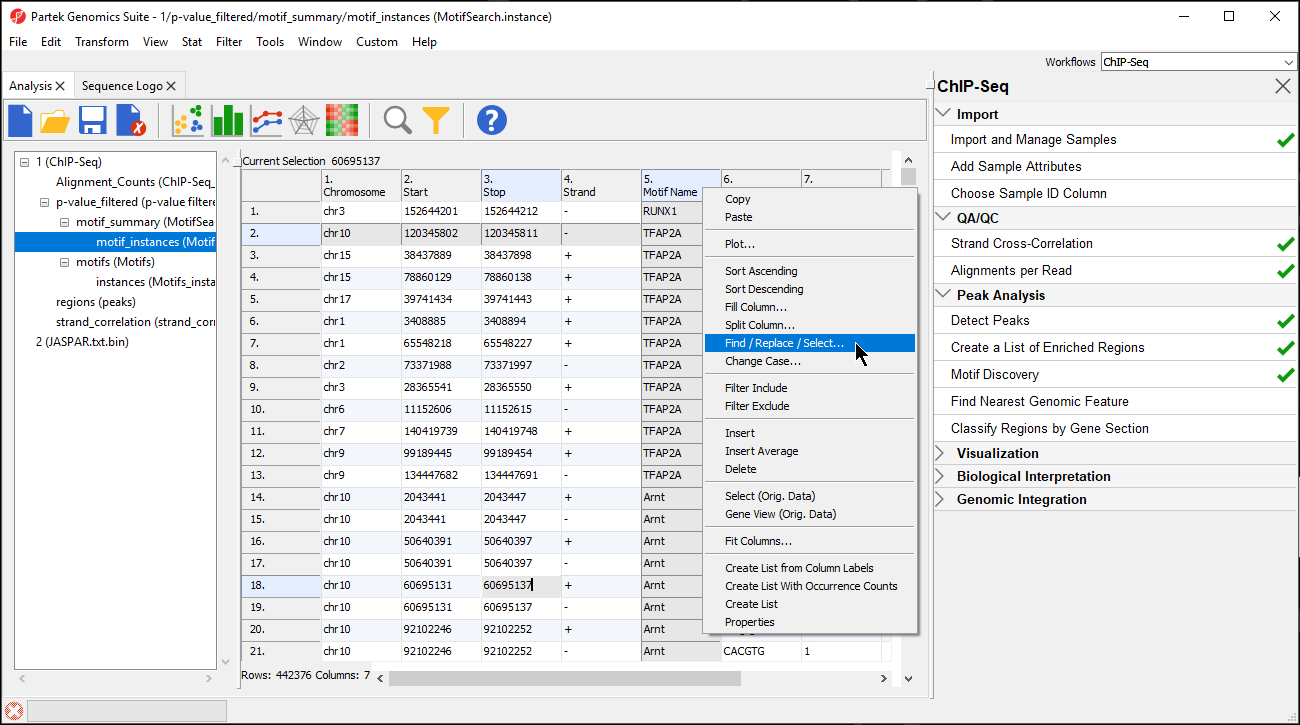
- Select the motif_instances spreadsheet in the spreadsheet tree
- Right-click the 5. Motif Name column
- Select Find / Replace / Select... from the pop-up menu (Figure 78)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
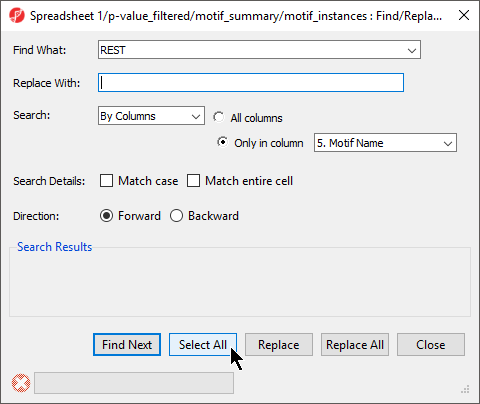
- Set Find What: to REST
- Select By Columns for Search:
- Select Only in column with 5. Motif Name selected form the drop-down menu
- Select Select All (Figure 89)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
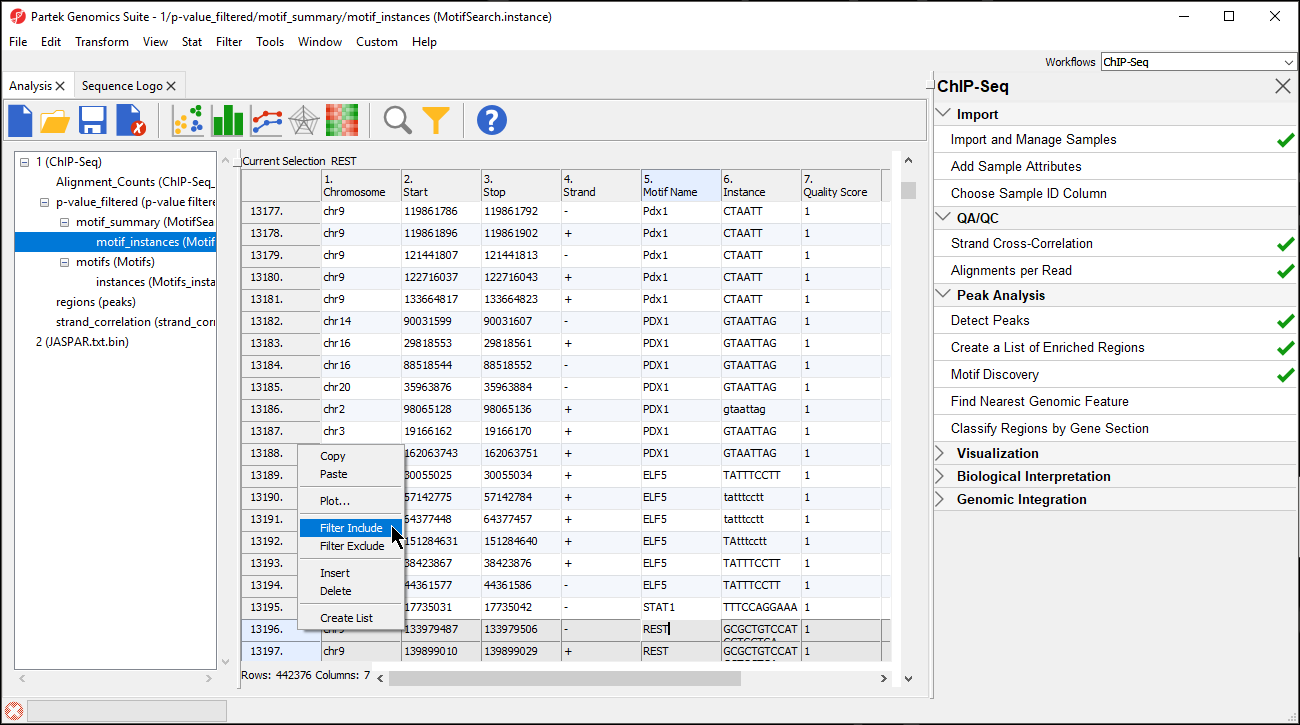
- Right-click on the first highlighted row visible; in this example, we see row 13196
- Select Filter Include from the pop-up menu (Figure 910)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The spreadsheet will now include 2098 rows and a black and yellow bar will appear on the right-hand side of the spreadsheet (Figure 1011). The black and yellow bar is a filter indicator showing the fraction of the spreadsheet currently visible as yellow and the filtered fraction as black.
...
Neuwald, A. F., Liu, J.S., & Lawrence, C.E. (1995). Gibbs motif sampling: detection of outer membrane repeats (Vol. 4). Protein Science.
| Page Turner | ||
|---|---|---|
|
| Additional assistance |
|---|
|
| Rate Macro | ||
|---|---|---|
|