Page History
When If your experimental design includes a sample or a group of sample serves samples serving as control baseline in the experiment design, other samples can be normalized a baseline control, you can normalize the experimental samples by subtracting or divided dividing by the baseline sample(s) , e.g. in PCR data using the Normalize to baseline task in Partek Flow. For example, in PCR experiments, the delta Ct values of control samples are subtracted from the delta Ct values of experimental samples to obtain delta-delta CT Ct values for the experimental samples.
The Normalize to baseline option is available at in the Normalization and Scaling section in of the context-sensitive menu (Figure 1) upon selection of any count matrix data node containing matrix of measurements of observations (e.g. samples) on features (e.g genes).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
There are three options to choose the baseline samples:
- use all samples
- use a group
- use matched pairs
Use all samples to create baseline
To normalize data to all the samples, choose to use calculate the baseline using the mean or median of all samples for each feature, and select choose to subtract baseline or ratio to baseline in for the normalization method (Figure 2), and click Finish.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Use a group of sample to create baseline
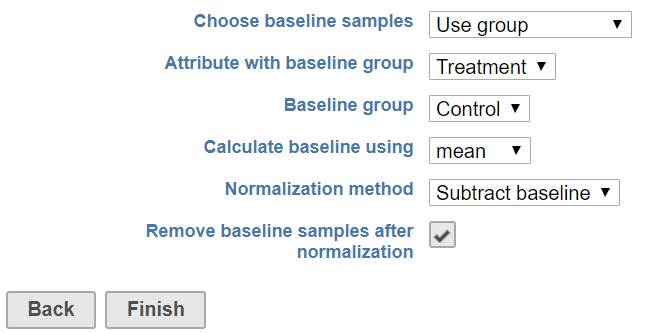
When there are is a subset of samples that serve as the baseline in the experiment, chooseselect use group as for Choose baseline samples. The specific group needs should be specified from the using sample attributes (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Choose this option use group, select the attribute containing the baseline group information, e.g. Treatment in this example, with the samples labeled as Control will be with the group Control for the Treatment attribute used as the baseline. After normalization, the control sample The control samples can be filtered out in the report after normalization by selecting the Remove baseline samples after normalization check buttonbox.
Use matched pairs
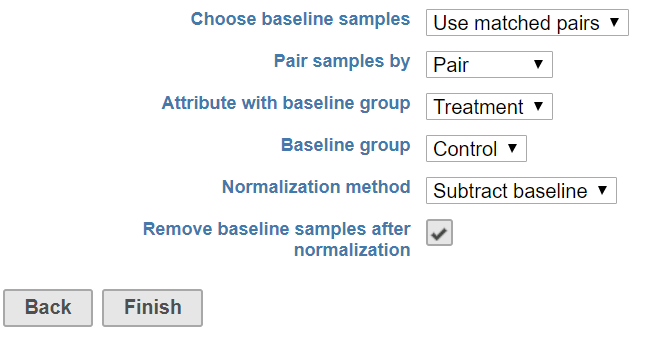
In a paired experiment design, when normalized one sample to its own control – typically there is only one sample serve as a control to its pair (Figure 4), an attribute on the pair information should be specified in addition to baseline group attribute.
When using matched pairs, one sample from each pair serves as the control. An attribute specifying the pairs must be selected in addition to an attribute designating which sample in each pair is the baseline sample (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
After normalization, all values for the control sample
...
will be either 0 or 1 depending
...
on the normalization method chosen, so we
...
recommend
...
removing baseline samples when using matched pairs.
The output of Normalize to baseline is a Normalized counts data node.
| Additional assistance |
|---|
...
| Rate Macro | ||
|---|---|---|
|