Page History
...
Space Ranger is a set of analysis pipelines that process Visium Spatial Gene Expression data with brightfield and fluorescence microscope images (1).
Space Ranger in Partek
...
Flow
Space Ranger 12.30.0 1 has been wrapped in Partek Flow as Space Ranger task. It does not include all the options and uses cases covered by Space Ranger, but The task takes .fastq and .jpeg/.tiff files as input and performs alignment, filtering, barcode counting, and UMI counting. The output is gene expression count matrix in a .h5 format (both raw and filtered are available for download via Task Details), which then becomes the starting point for downstream analysis in Partek Flow.as well as a .zip file with spatial files (image). Note that the Space Ranger task in Partek Flow does not include all the options and uses cases covered by the Space Ranger pipelines of 10x Genomics.
Note: when using the Space Ranger task in Partek Flow, there are more restrictions on the sample name-- sample name can only contain letters, digits, underscores and dashes. Please Edit the sample names on Data tab in Partek Flow to remove any other characters, e.g. space etc.
Running Space Ranger in Partek Flow
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
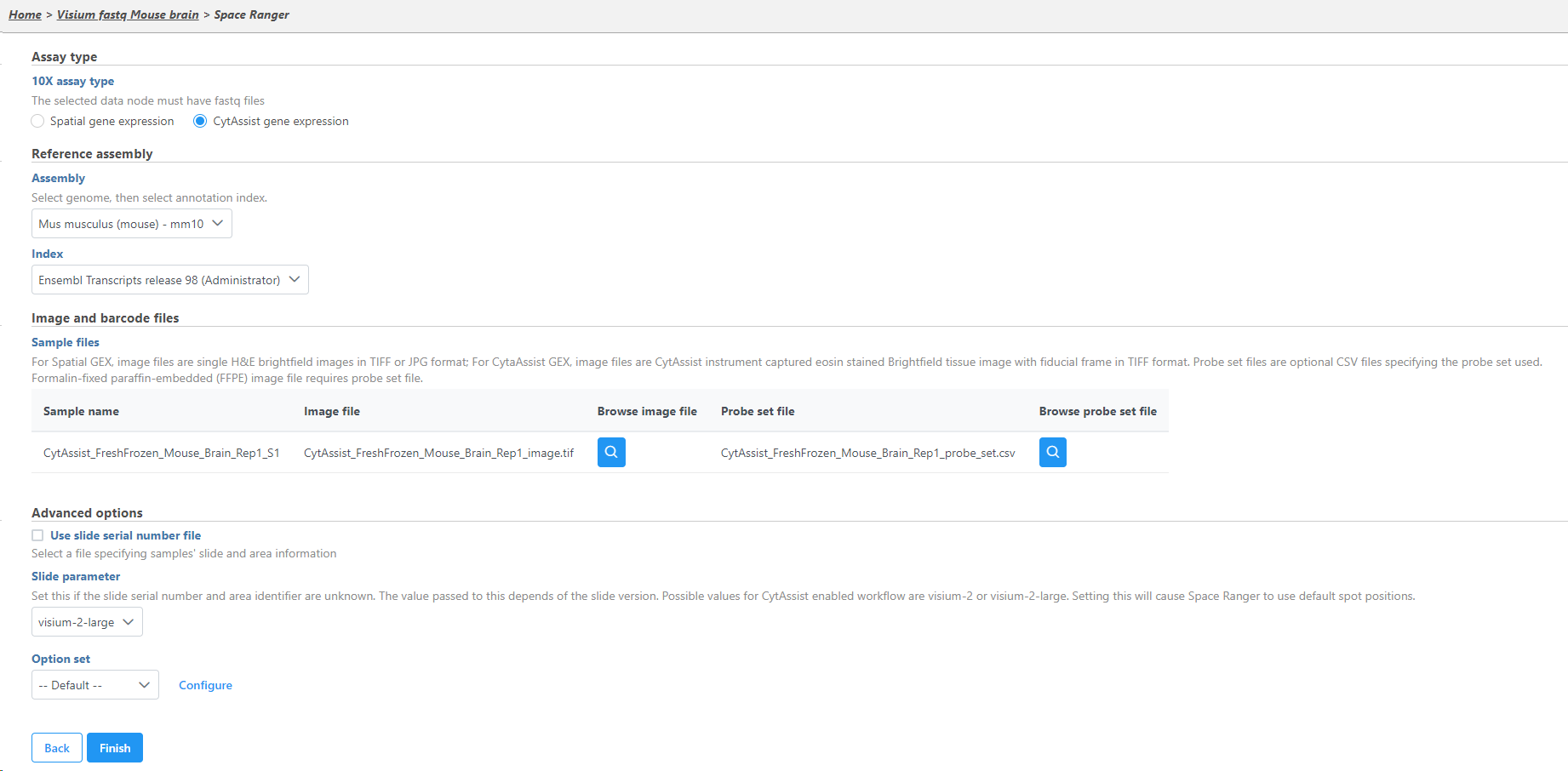
If this is the first time Select the 10x assay type. Choose CytAssist gene expression if you are using the Visium CytAssist gene expression library.
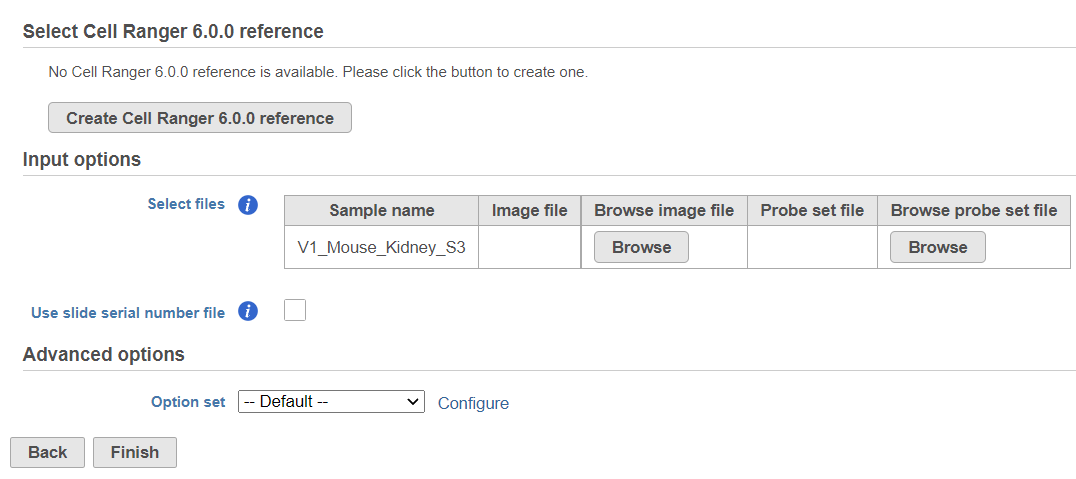
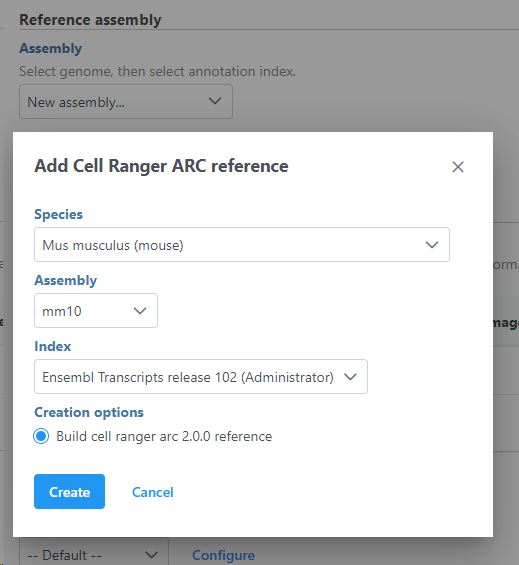
For both Space Ranger or Cell Ranger, you will need to create a Cell Ranger reference tasks, a Reference assembly is required (Figure 2). To define the Reference assembly, first select the Genome build for the organism of interest, then select the annotation Index. In this manner, custom libraries can be created (e.g. keep the Genome build but change the annotation Index).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
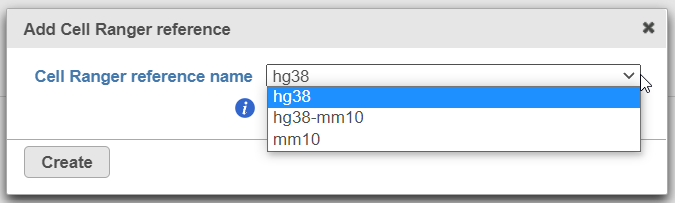
To create add a new reference, push the Create Cell Ranger 6.0.0 reference button and pick the appropriate Cell Ranger reference name new species genome, choose New assembly from the drop down for Genome build which will open a new window with configuration options to edit, then click Create (Figure 3) and then push Create.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Downloading a reference is a separate task and may take some time. However, it will become immediately available in the Cell Ranger 6.0.0. reference drop down (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Alternatively, Cell Ranger reference can be added beforehand, to the Library file management page (on the Other library files tab). At the moment, the following references are available for download from Partek: hg38, mm10, and hg38-mm10 (chimaeric genome).
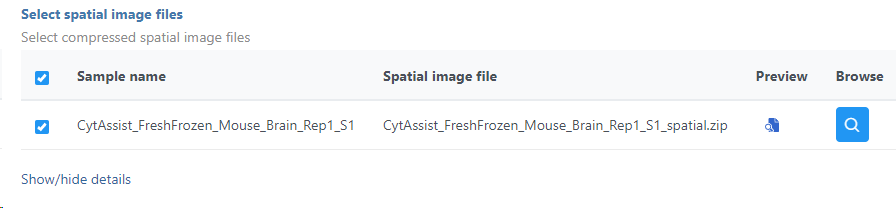
The sample table under Input options has one row per sample (Figure 2). Image file is required, and that is a single hematoxylin and eosin brightfield image in either .jpg or .tiff format. Click on the Browse button under Browse image file and the file browser will come up. Point to the image file and push Continue. Formalin-fixed paraffin-embedded (FFPE) image files require the Probe set file otherwise it is optional; it is a .csv file specifying the probe set used (=target panel).
If you want to specify the sample's slide and area information, tick mark the check the box by the Use slide serial number file under Advanced options and then click Browse to point to the file. The file should be tab-delimited with samples on rows. The first column is the sample name, the slide name is on the second column, slide area is on the third column.Click
If the Slide serial number is not available for CytAssist samples, the Slide parameter should be specified where visium-2 corresponds to a 6.5 x 6.5 mm capture area and visium-2-large corresponds to a 11 x 11 mm capture area.
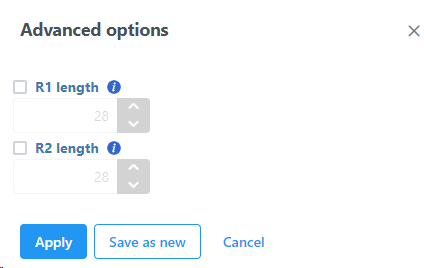
If necessary, click on the Configure link in the Advanced options section to open the Advanced options Options dialog (Figure 54). Use R1 length to hard trim the R1 reads to specified length; use R2 length to hard trim the R2 reads to specified length. Use the Memory limit option to cap the RAM usage (in GB).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Space Ranger task report
...
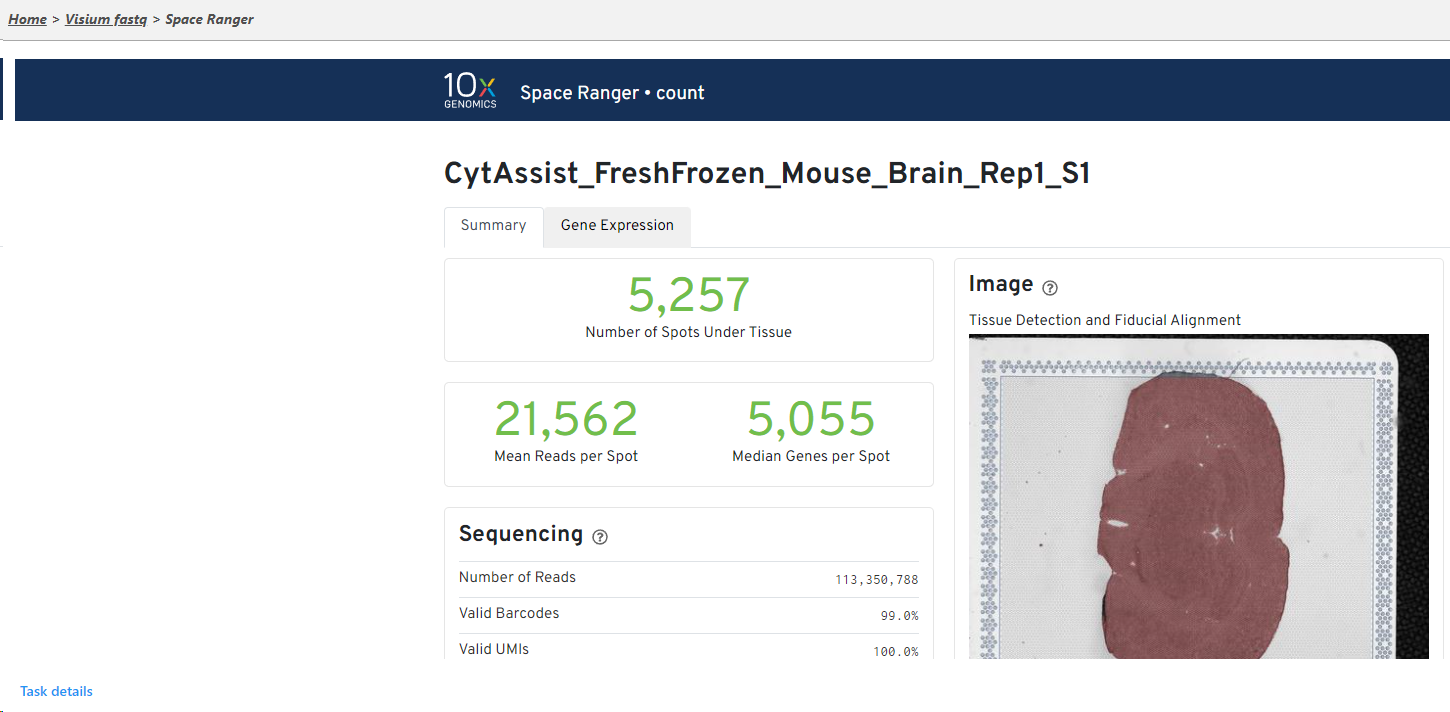
The result of Space Ranger task is the Single cell counts data node, which contains the gene expression data. Double click the Single cell counts node to invoke the report the task report (Figure 65) which is the same as the ‘Summary HTML’ from Space Ranger outputthe original 10x Genomic pipelines. Task report is sample based. You can use the dropdown list in top left to switch samples (not shown in Figure 6). Each report consists of two pages: Summary and Analysis. For deatailsdetails, please visit 10x Genomics web page.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Annotate Visium image
After creating the Single cell counts node, the next step is to associate the microscopy image with the expression data. To start, select the Single cell counts data node and then go to Annotate Visium image in the toolbox (Figure 76).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The setup page shows the sample table (one sample per row; Figure 87). Click on the Browse button to open the file browser and point to the file <project-name>_spatial.zip, created by the Space Ranger task. After that, click on Finish to launch the Annotate task.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
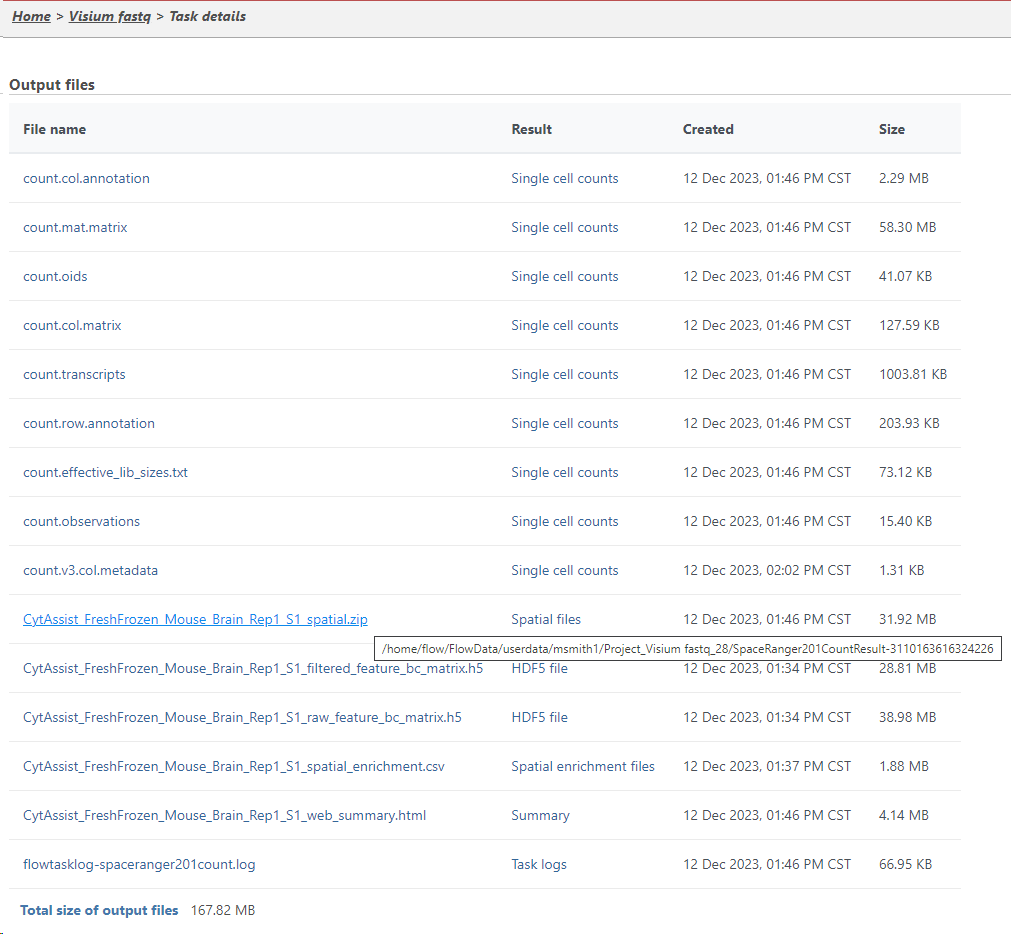
You can find the location of the <project-name>_spatial.zip file using the following sepssteps. Selcte Select the Space Ranger task node (i.e. the rectangle) and then click on the Task Details (toolbox). Click on the Output files link to open the page with the list of files created by the Space Ranger task. Mouse over any of the files to see the directory in which the file is located. Figure 9 8 shows the path to the .zip file which is required for Annotate Visium image.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new data node, Annotated counts, will be generated (Figure 9).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The Annotated counts node is Split by sample. This means that any tasks performed from this node will also be split by sample. Invoke tasks from the Single cell counts node to combine samples for analyses.
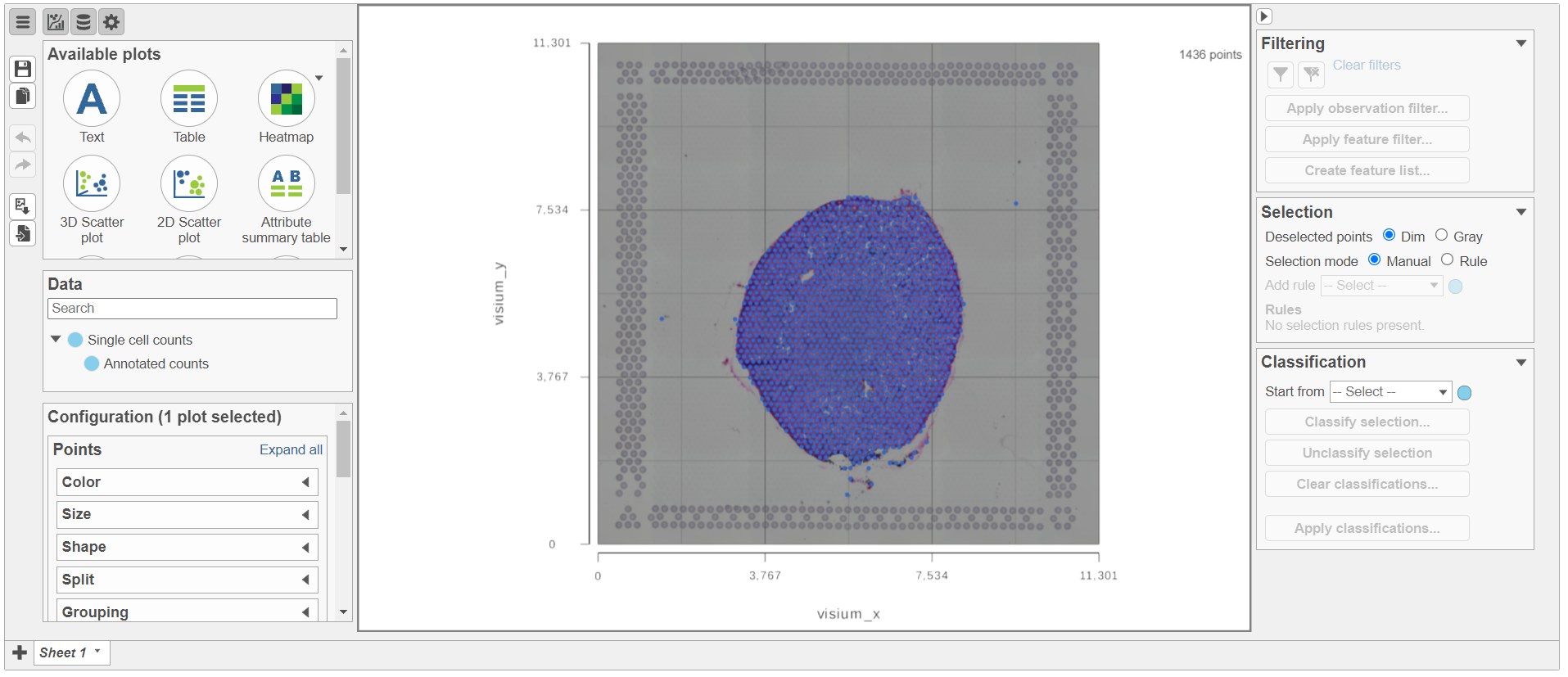
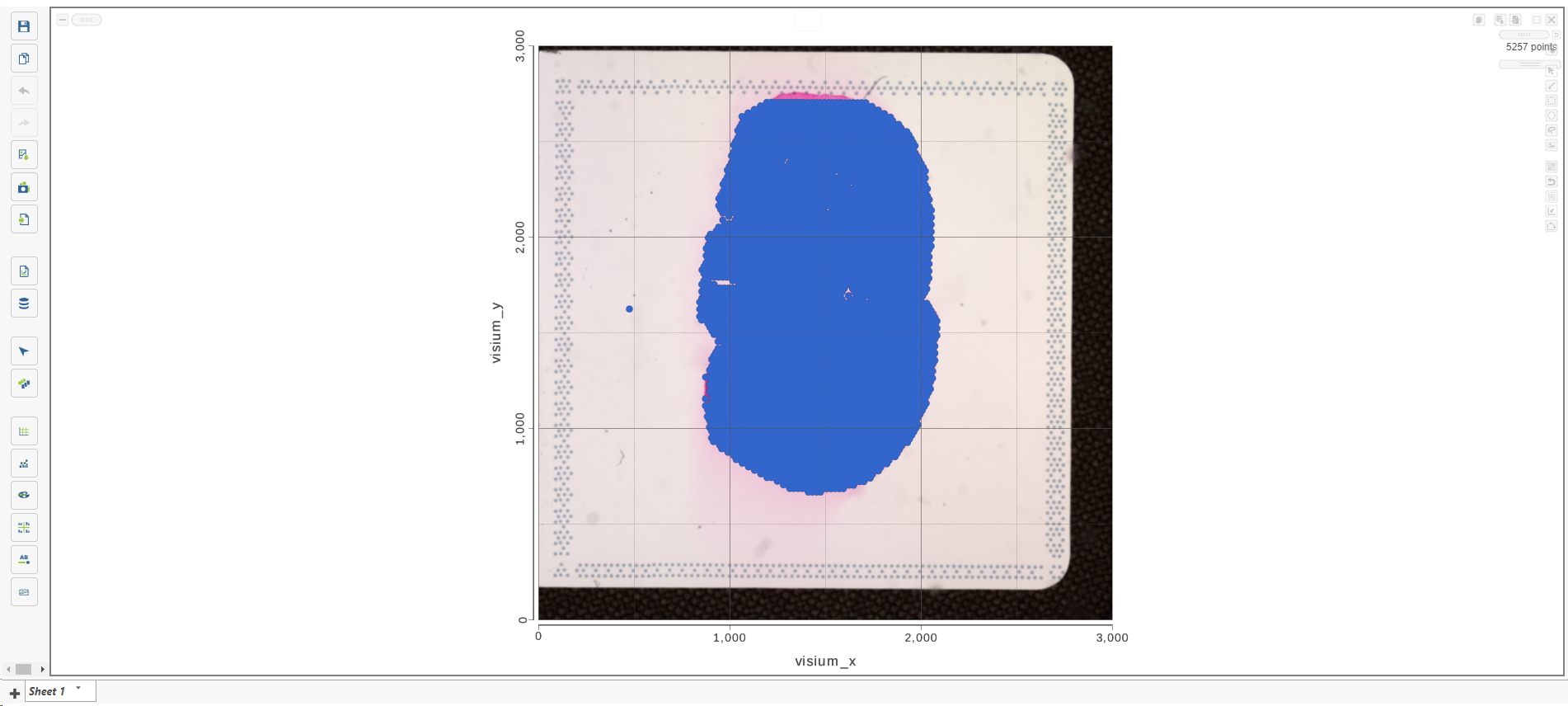
Annotate Visium image task creates a new node, Annotated counts. Double click on the Annotated counts node to invoke the Data Viewer showing data points overlaid on top of the microscopy image (Figure 10).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||