A histogram is a plot that summarizes the underlying frequency of a set of data with the variable of interest on one axis and the frequency distribution of that variable in the other axis. In Partek Flow, histogram can be invoked on continuous or categorical variable.
Invoking a histogram
From a data viewer session, drag the histogram icon onto the data viewer canvas click on New plot > Bar chart (Figure 1).
| Numbered figure captions |
|---|
| SubtitleText | Drag histogram plot (red rectangle) onto data viewer canvassSelect the Bar chart option from the New plot menu. |
|---|
| AnchorName | Invoke Histogram 1 |
|---|
|
 Image Removed Image Removed Image Added Image Added
|
Upon dropping the histogram clinking on the canvas Bar chart menu, a dialogue opens up with the different data nodes sources that can be displayed on the histogram. Select your data node of interest and the content data (Figure 2).
| Numbered figure captions |
|---|
| SubtitleText | Select Normalized count the appropriate data node (red) and content data to display on the histogram plot |
|---|
| AnchorName | Plot datanode on histogram |
|---|
|
 Image Removed Image Removed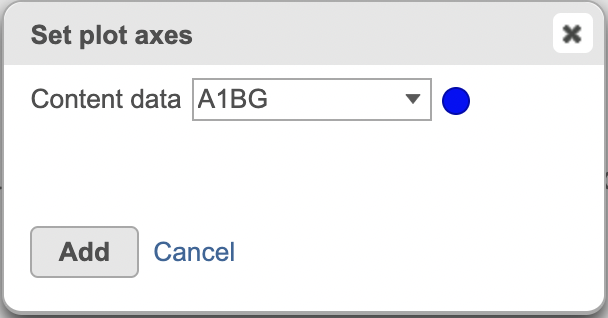 Image Added Image Added
|
The first row in the data will be displayed by default in the histogram and in this case, it is the histogram of the expression values for the gene A1BG (Figure 3).
...
| Numbered figure captions |
|---|
| SubtitleText | Histogram showing the distribution of AIBG expression (red) |
|---|
| AnchorName | Default display on histogram |
|---|
|
 Image Removed Image Removed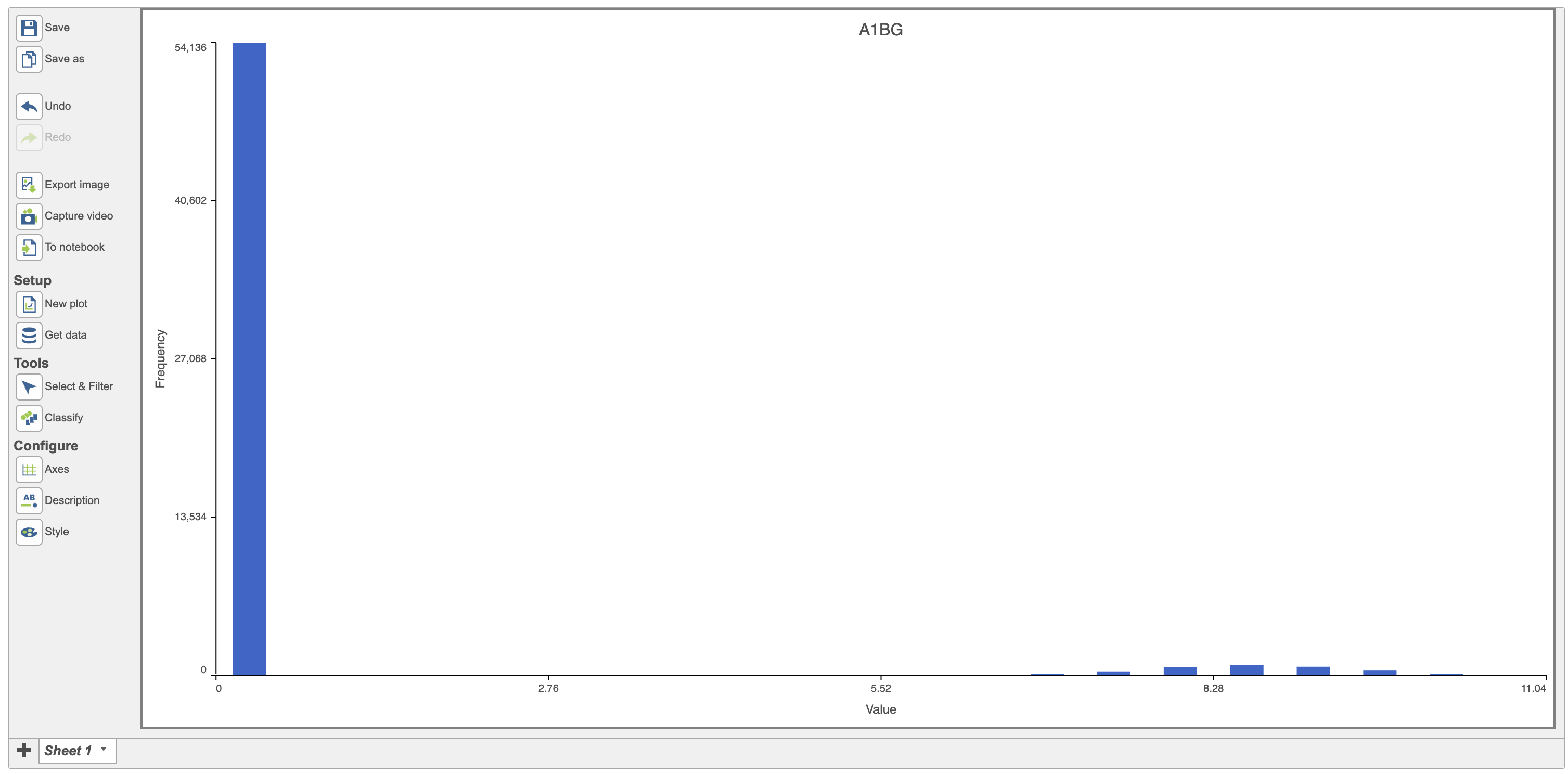 Image Added Image Added
|
Histogram of Continuous variables
Change the data displayed on the histogram by using the Configuration settings > Content > Data and Configure > Axes menu and selecting the desired variable to display. Here the data displayed was switched to “Expressed genes” which is a continuous variable (Figure 4).
...
| Numbered figure captions |
|---|
| SubtitleText | Histogram showing the distribution of expressed genes variable (red rectangle) |
|---|
| AnchorName | Histogram of expressed genes |
|---|
|
 Image Removed Image Removed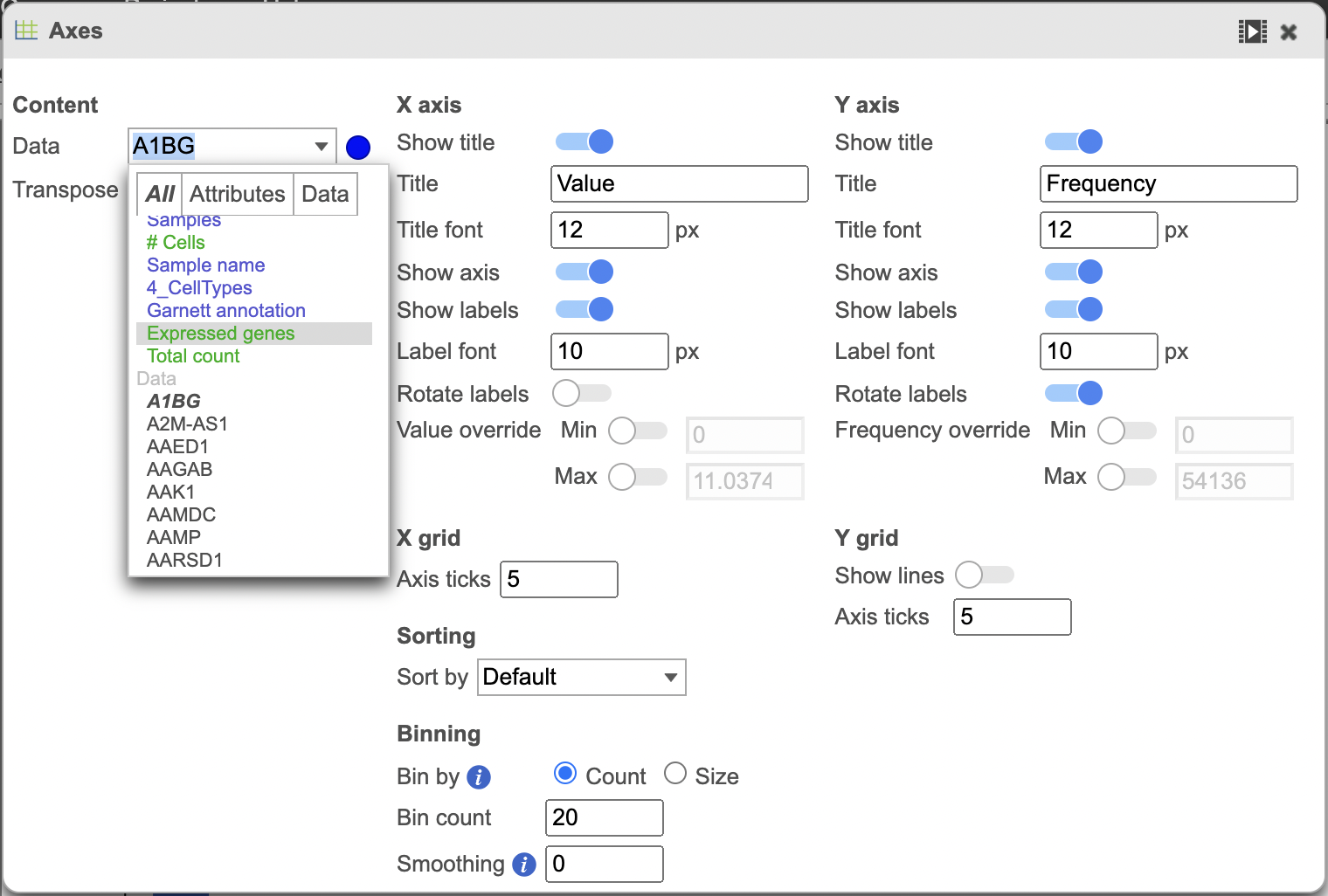 Image Added Image Added
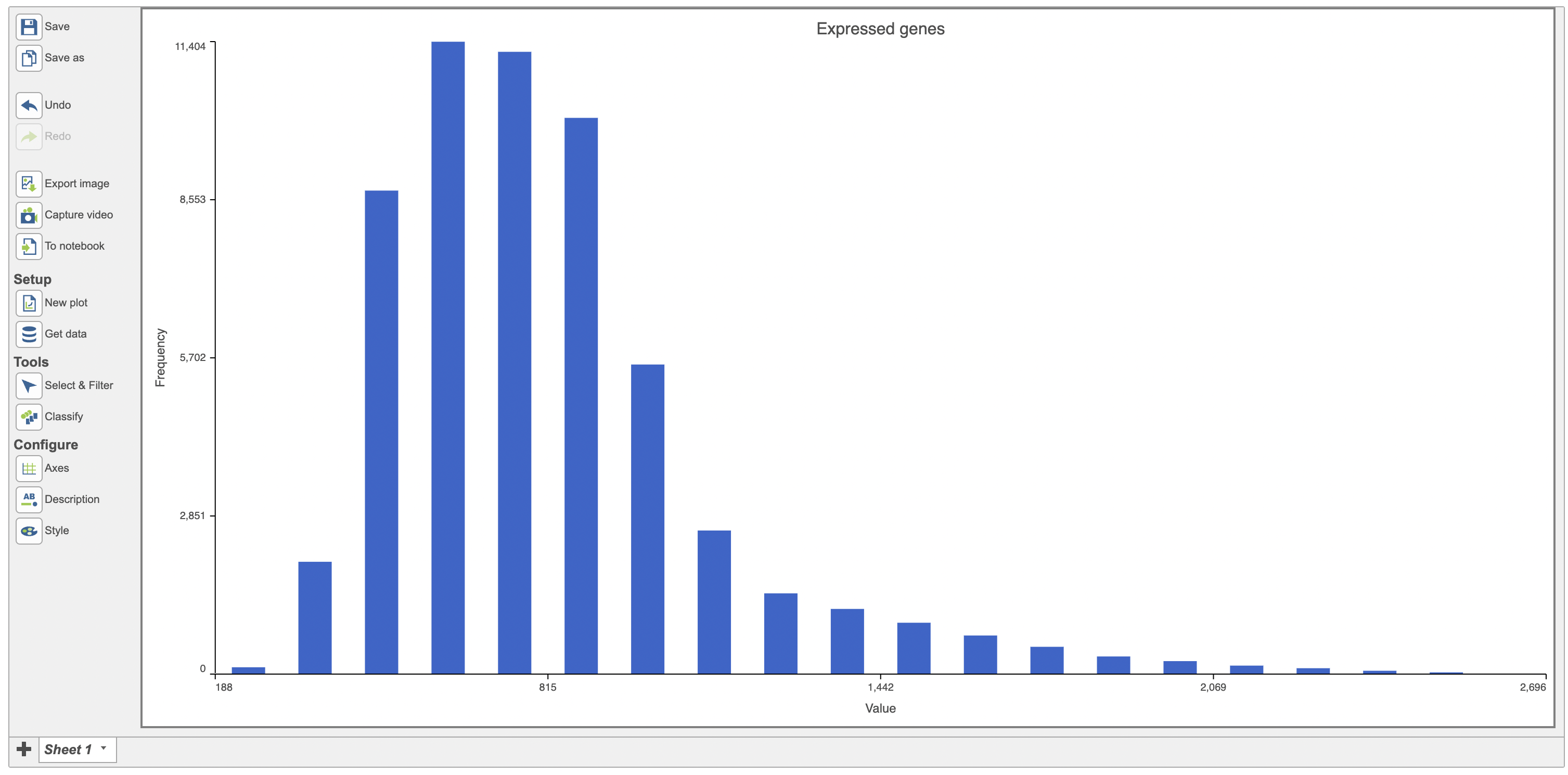 Image Added Image Added
|
Use the "Sort by" function to sort the plot. The default sorting is by Value on the x-axis and this default setting is sorted in ascending order. Users have the option to change that by changing the Default to value or frequency in the sort option (Figure 5)
...
| Numbered figure captions |
|---|
| SubtitleText | Sort by function can be by Value or Frequency (red) |
|---|
| AnchorName | Sorting options for continuous variable |
|---|
|
 Image Removed Image Removed
|
...
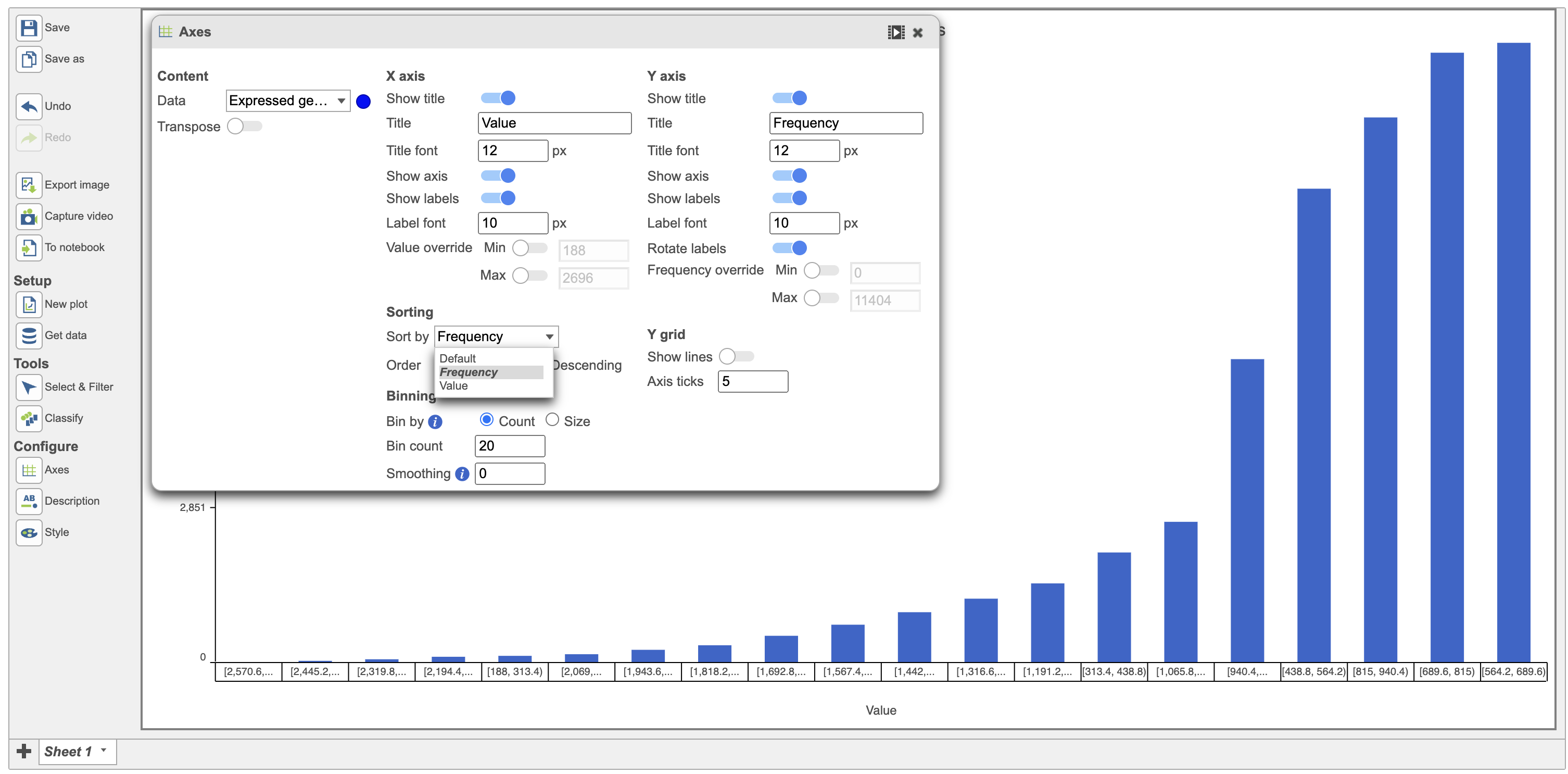 Image Added Image Added
|
Users can color the histograms by a categorical attribute using the Color by function (in red below). The bars were colored by the graph-based classifications in the example below (Figure 6).
| Numbered figure captions |
|---|
| SubtitleText | Histogram of expressed genes sorted by Value in descending order (red) |
|---|
| AnchorName | Image of expressed genes sorted by Value and in descending order |
|---|
|
 Image Removed Image Removed
|
| annotated by automatic classifications | | AnchorName | Annotate histogram |
|---|
|
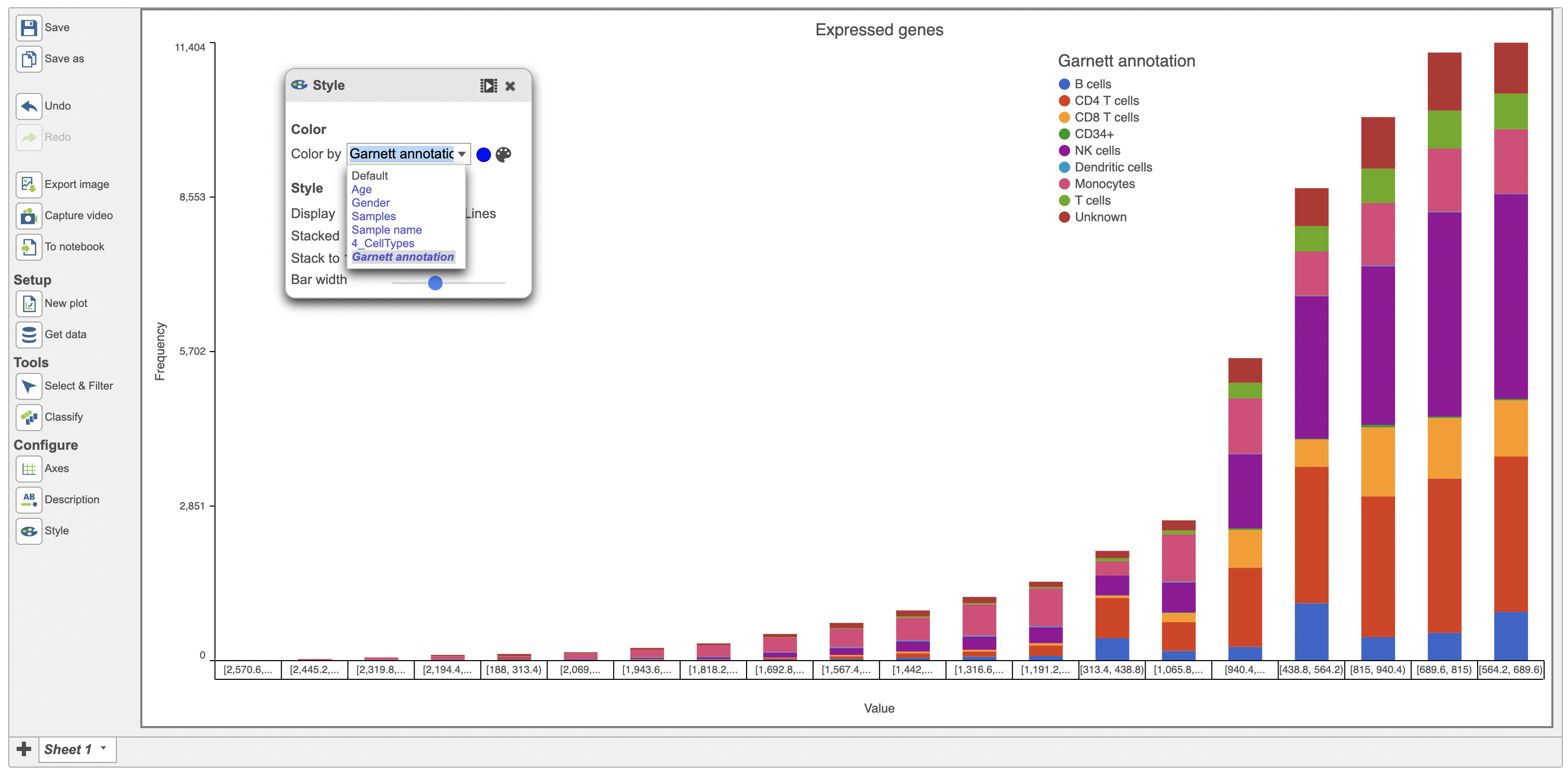 Image Added Image Added
|
The bars in the histogram above were stacked. They can be unstacked using the Style menu as seen below in red (Figure 7).
| Numbered figure captions |
|---|
| SubtitleText | Unstacked bars in the histogram plot |
|---|
| AnchorName | Unstacking histogram |
|---|
|
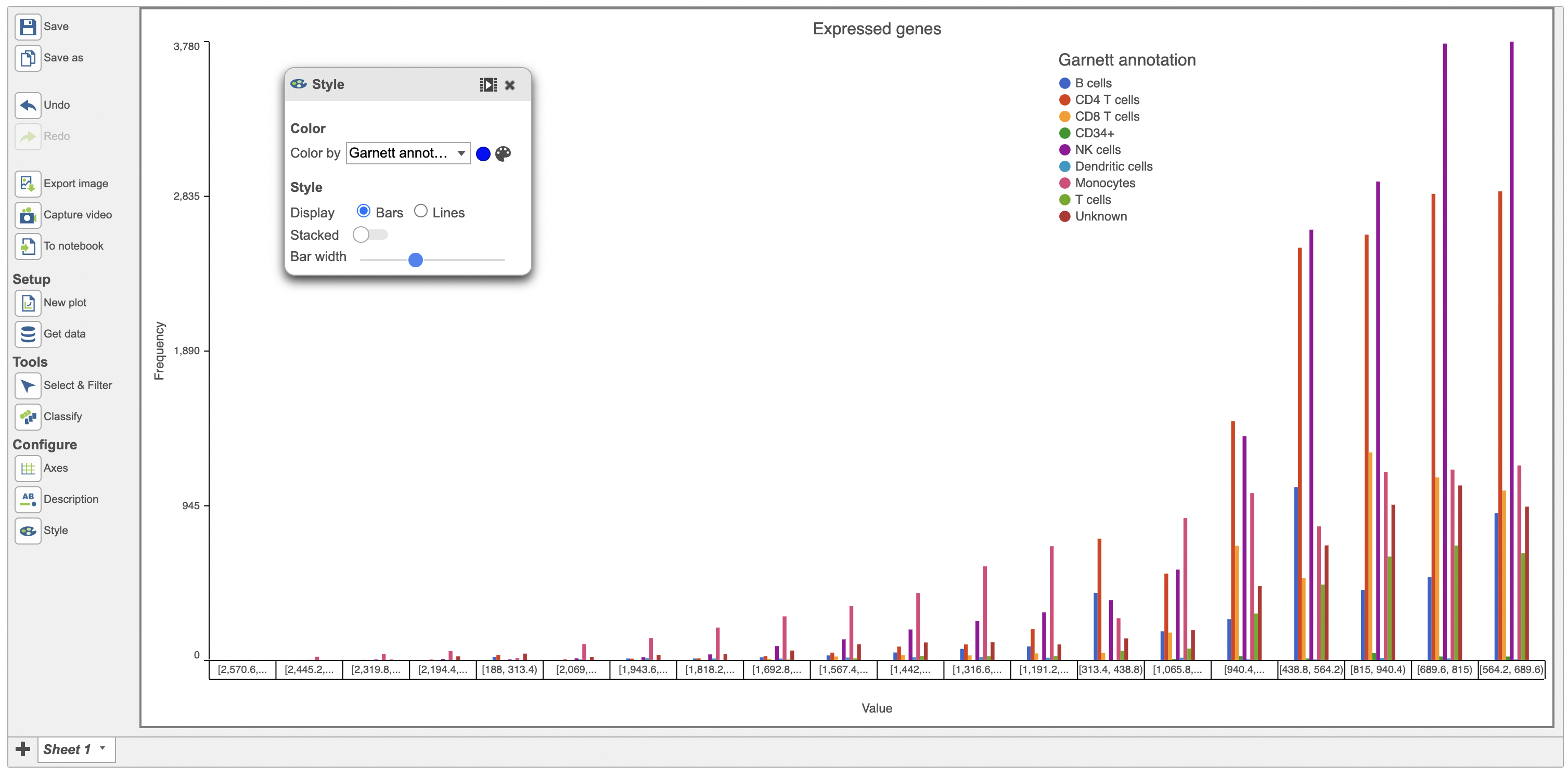 Image Added Image Added
|
Users also have the option to bin by either Count or Size. When binned by Count, the user specifies the number of bins for the data and the distribution is fit into the specified number of bins. Data below is binned by Count (Figure 78).
| Numbered figure captions |
|---|
| SubtitleText | Histogram of expressed genes with number of bins specified as 510 |
|---|
| AnchorName | Image of expressed genes with bin Count of 5 |
|---|
|
 Image Removed Image Removed
|
| Numbered figure captions |
|---|
| SubtitleText | The mouseover example Pie chart. |
|---|
| AnchorName | The mouseover Pie chart |
|---|
|
 Image Removed Image Removed
|
Configuration card (red rectangle in Figure 6) for Pie chart in Flow includes the options:
- Data: multiple categorical attributes can be added to data source; Users are allowed to rearrange their order by dragging when having multiple categorical attributes (Figure 7).
- Split by: split the current Pie chart by a second categorical attribute (Figure 6)
- Color mode: Unique colors (default), Similar colors.
- Title: Title name, Title font size (16 px as default)
- Style: Order slices by Count or Category
| Numbered figure captions |
|---|
| SubtitleText | Configuration and splitted example of Pie chart. |
|---|
| AnchorName | Configuration and splitted example of Pie chart |
|---|
|
 Image Removed Image Removed
|
...
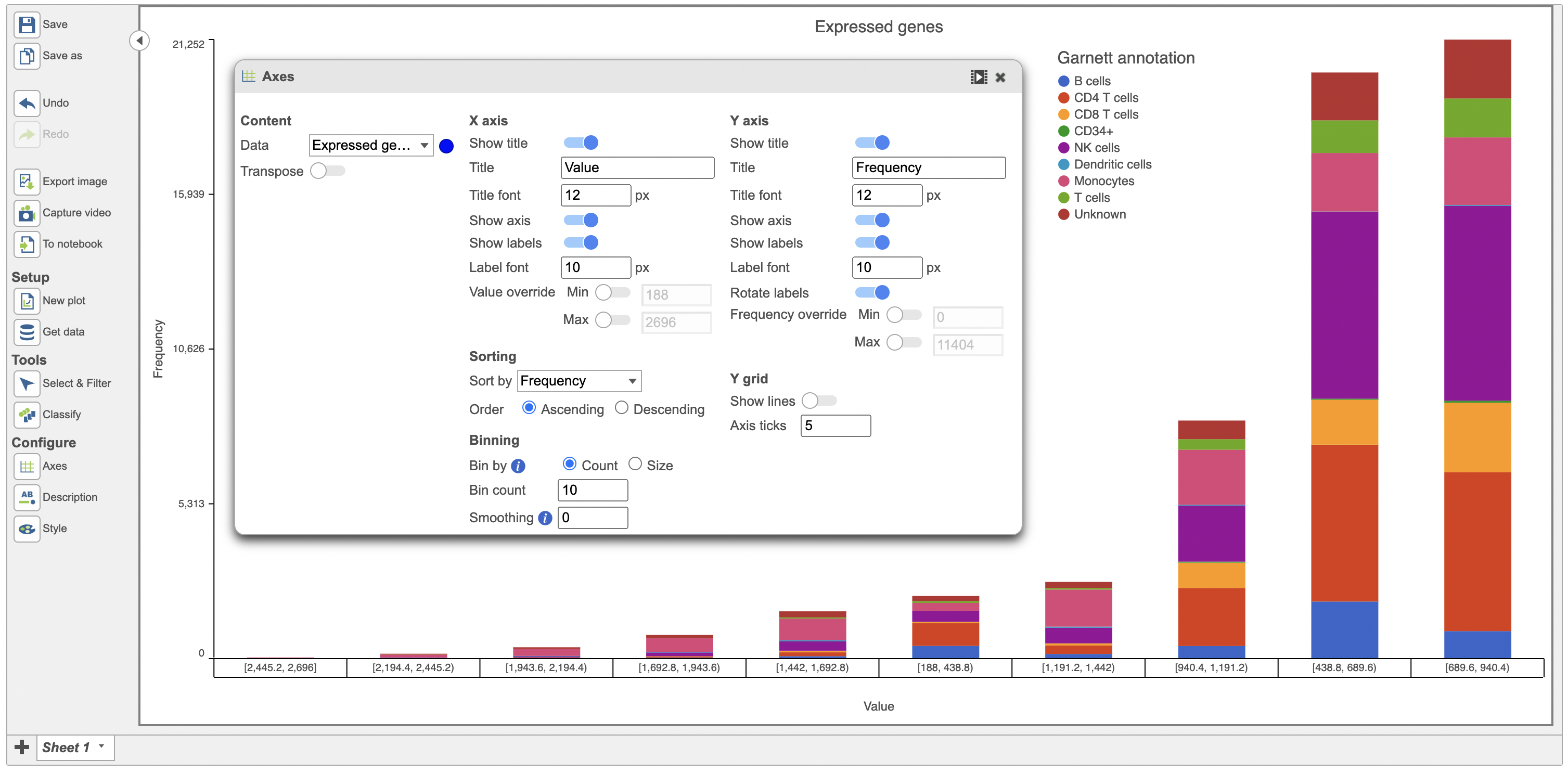 Image Added Image Added
|
When binned by Size, the user specifies the number of items in the bin (size of a bin). This is used to calculate the number of bins required for the data. Data below is binned by Size (Figure 10).
| Numbered figure captions |
|---|
| SubtitleText | Different modes of Pie Chart. |
|---|
| AnchorName | Different modes of Pie Chart |
|---|
|
 Image Removed Image Removed
|
Once you are pleased with the appearance of the Pie plot, push Save image button to save it to the local machine or click Save button to save the Data Viewer. The resulting dialog (Figure 8) controls the Format,Size and Resolution of the image file. The image will be saved in your favorite format (.svg, .png and .pdf).
| Numbered figure captions |
|---|
| SubtitleText | Save image dialog (default settings) |
|---|
| AnchorName | Save image |
|---|
|
 Image Removed Image Removed
|
| Histogram of expressed genes with size of bin specified as 75 | | AnchorName | Value binned by Size image |
|---|
|
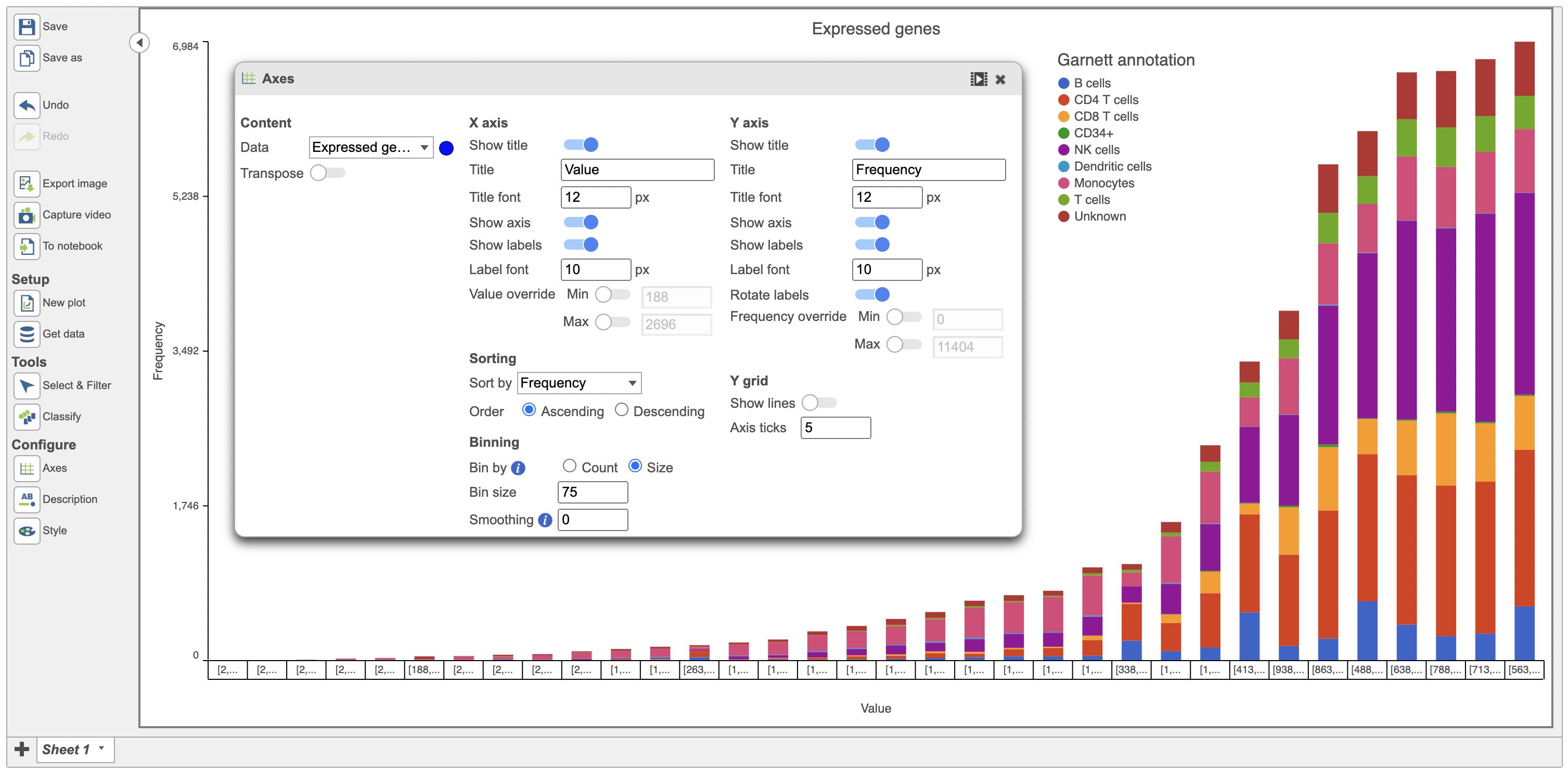 Image Added Image Added
|
ghjkl
Click the Save image button  Image Added to save a PNG or SVG image to your computer.
Image Added to save a PNG or SVG image to your computer.
Click the Send to notebook button  Image Added to send the image to a page in the Notebook.
Image Added to send the image to a page in the Notebook.
ghjkl
ghjkl
ghjkl
Additional Assistance
If you need additional assistance, please visit our support page to submit a help ticket or find phone numbers for regional support.
...




















