Page History
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
How to import a study from GEO / ENA
Partek® Flow® supports single cell data analysis in count matrix text format. Each matrix text file is assumed to represent on sample, each value in the matrix represents expression value of a feature (e.g. a gene, or a transcript) in a cell. The expression value can be raw count, or normalized count. The requirment of the format of each text file should be the same as count matrix data.
To import a sample during import data, choose Import single cell data option (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Specify text file location, only one text file (in other words one sample) can be imported at once, preview of the file will be displayed, configuration of the file format is the same as Import count matrix data. In addition, you need to specify Sample information and Gene deduplication method (Figure 2).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Click Finish, the sample will be imported, on the data tab, number of cells in the sample will be displayed.
To import multiple samples, repeat the above steps by clicking Import data on the data tab
Imported projects from GEO / ENA
Common Issues
FAQ
If a project is publicly available in the Gene Expression Omnibus (GEO) and European Nucleotide Archive (ENA) databases, you can import associated FASTQ files and sample attributes automatically into Partek Flow.
- On the Homepage click New Project to create a project and give the project a name
- Click Add data
- Select fastq as the file type after choosing between Single cell or Bulk as the assay types
- Click Next
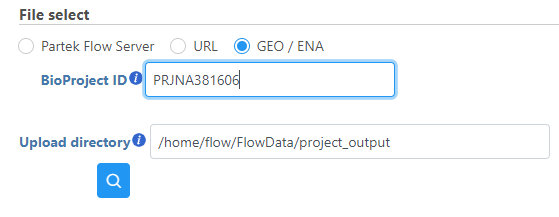
- Choose GEO / ENA
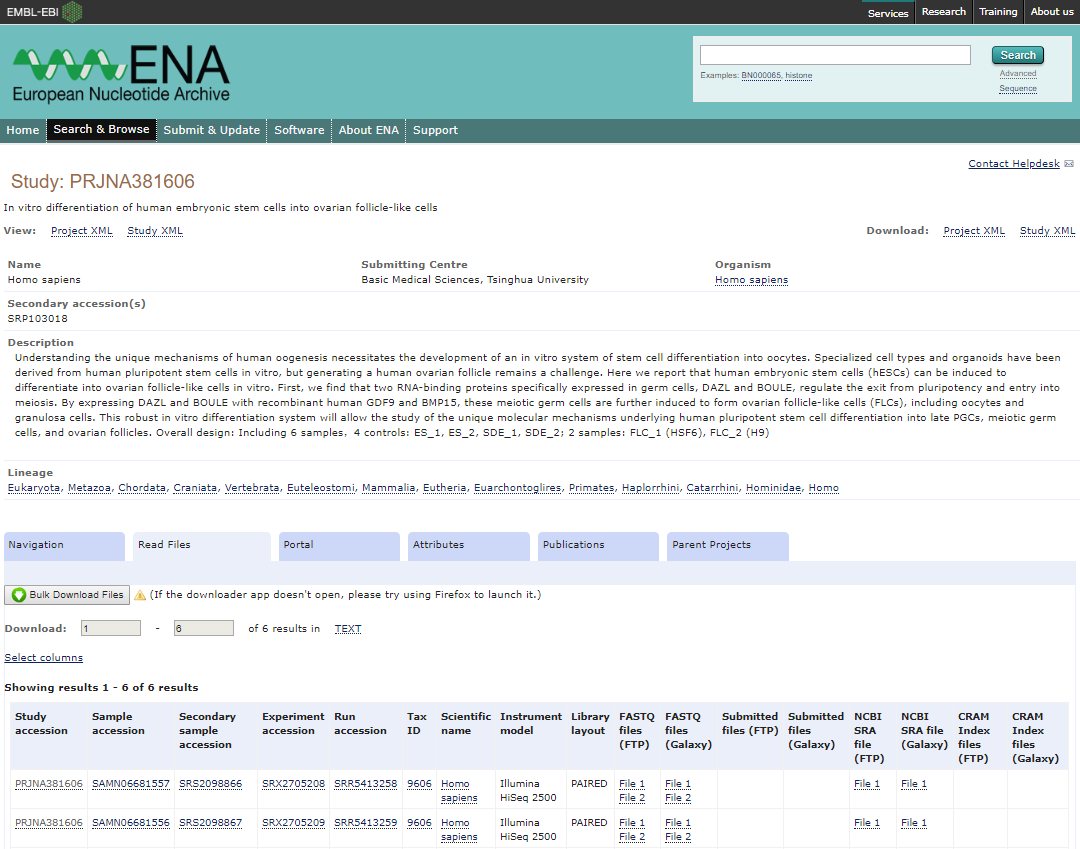
- Enter the BioProject ID of the data set you would like to download. The format of a BioProject ID is PRJNA followed by one to six numbers (e.g. PRJNA381606)
A GEO ID can also be used in the format GSE followed by one to five numbers (e.g. GSE71578).
- Click Finish
It may take a while for the download to complete depending on the size of the data. FASTQ files are downloaded from the ENA BioProject page.
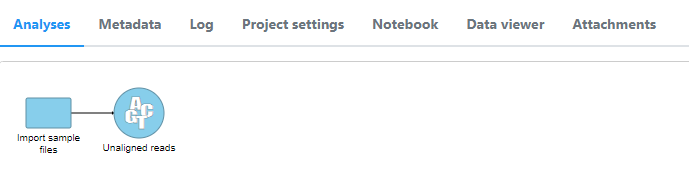
- FASTQ files will be added as an Unaligned reads data node in the Analyses tab
Common Issues
Error Message - The project did not yield any data. Double-check the project ID, or try importing the data manually
If the study is not publicly available in both GEO and ENA, project import will not succeed.
The project was imported, but the Analyses tab is empty and there are no FASTQ files
If there is an ENA project, but the FASTQ files are not available through ENA, the project will be created, but data will not be imported.
Something is missing or the import failed
A variety of other issues and irregularities can cause imports to not succeed or partially succeed, including, but not limited to, a BioProject having multiple associated GSE IDs, incomplete information on the GEO or ENA page, and either the GEO or ENA project not being publicly available.
FAQ
What are GEO and ENA?
The Gene Expression Omnibus (GEO) and the European Nucleotide Archive (ENA) are web-accessible public repositories for genomic data and experiments. Access and learn more about their resources at their respective websites:
GEO - https://www.ncbi.nlm.nih.gov/geo/
ENA - https://www.ebi.ac.uk/ena
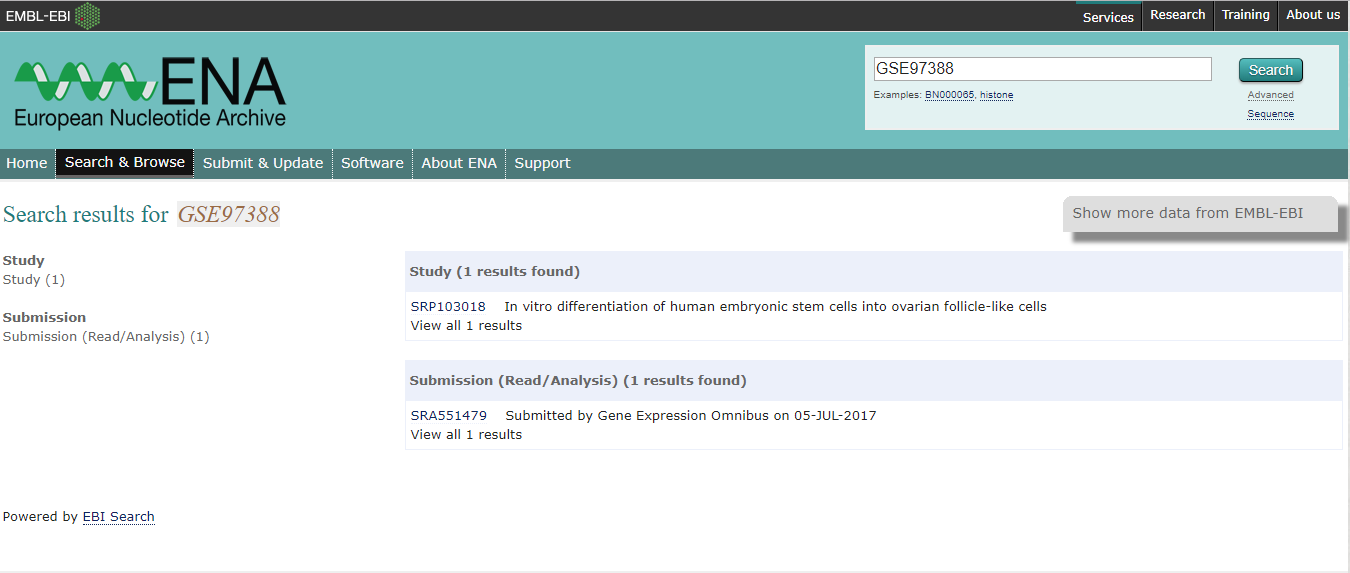
How do I know if a GEO project is also in ENA?
- You can search ENA using the GEO ID (e.g., GSE71578) to check if there is a matching ENA project.
- Open the Study result to view the BioProject ID (e.g., PRJNA381606) and a table with information about the samples and files included in the project
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...