Page History
Gene Set Enrichment Analysis GSEA is a bioinformatics tool that determines whether a set of genes (e.g. a gene ontology (GO) group or a pathway) shows statistically significant, concordant differences between two experimental groups (1,2). Briefly, the goal of GSEA is to determine whether the genes belonging to a gene set are randomly distributed throughout the ranked (by expression) list of all the genes that should be taken into consideration (e.g. gene model), or are primarily found at the top or at the bottom of the list.
| Table of Contents |
|---|
Prerequisites
To run GSEA, your project has to contain at least one categorical factor with exactly two levels (e.g. Treated and Control). If you are running GSEA on RNA-seq data, note that some common normalisation transformations, such as fragments/reads per kilobase of transcript per million mapped reads (FPKM/RPKM) or transcripts per million (TPM) are not considered suitable for GSEA (for more information, please see GSEA documentation). Instead, you should use an approach such as DESeq2 normalisation, trimmed means of M (TMM), or geometric mean.
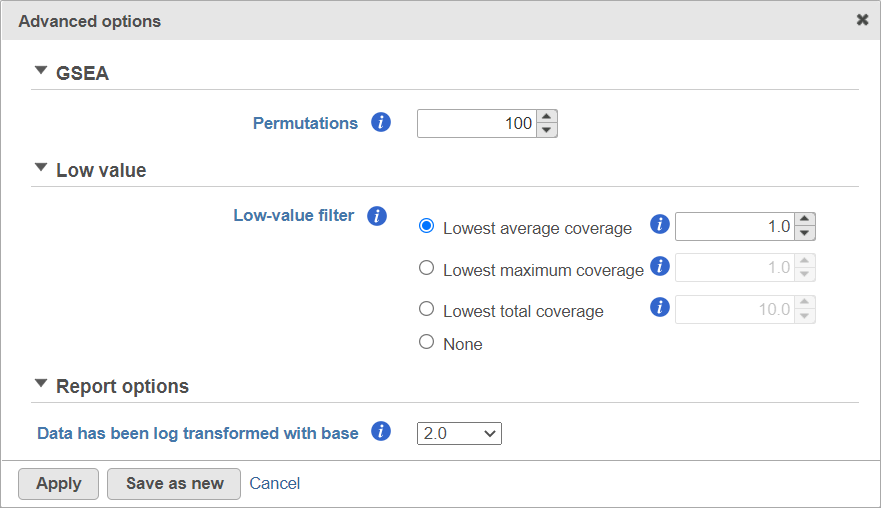
Running GSEA
To launch GSEA, select the data node with normalised data and then go to Biological interpretation > GSEA (Figure 1).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
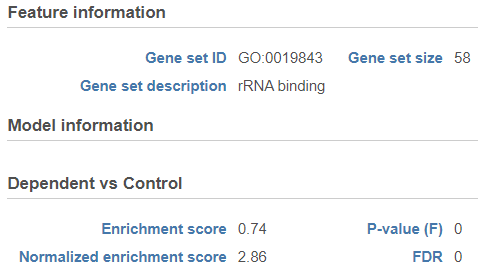
GSEA Results
When the task completes, double click on the GSEA task node (Figure 8) to view the report.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
References
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545-15550. doi:10.1073/pnas.0506580102
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267-273. doi:10.1038/ng1180