Page History
...
In order to know whether the data has been adapter-trimmed for microRNA data, we can look at the pre-alignment QA/QC of the raw data, specifically the read length distribution. If the read length distribution peaks at approximately 22-23 bases, this usually means the data has been adapter-trimmed. However, if you have a fixed length distribution, then very likely the data is not adapter-trimmed and you will need to get the adapter sequence from your vendor or service provider and use the Trim adapter function to trim away the adapter sequence.
Partek® Flow® software Partek Flow software wraps Cutadapt [1], a widely used tool for adapter trimming. It can be used to trim adapter sequences in nucleotide-space data as well as color-space data.
...
After that, the best adapter will be chosen based on the number of matching bases to the read. If there is a tie, adapters of the same type will be chosen in the order they are provided and adapters of different types will be chosen by type in the following order: first 3', then 5' or 3', and lastly 5' adapters.
Advanced Options for Trim Adapters
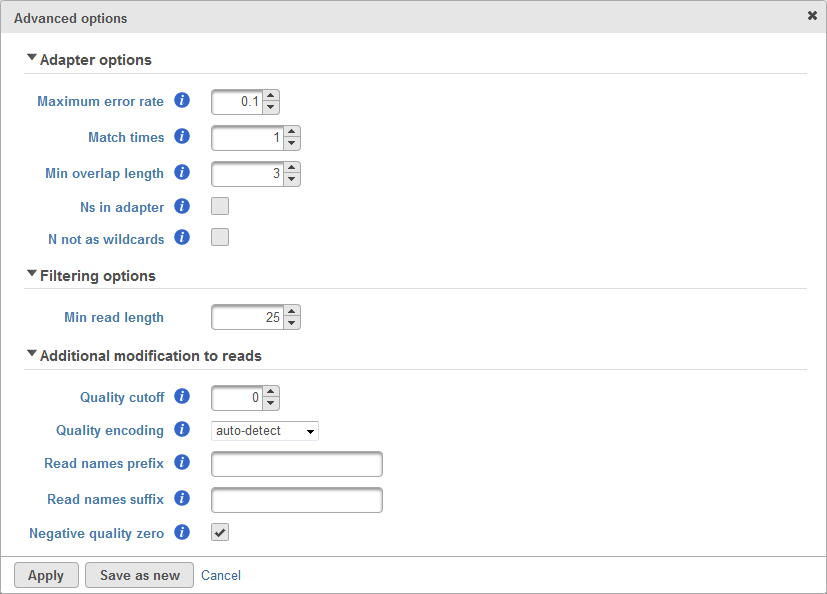
There are cases when the Trim adapters function does not work properly, for example: the existence of N's base in the read, etc. Therefore, there are advanced options which allows user to configure how the matching is done to trim adapter sequence. The advanced options dialog box is shown in Figure 2.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
The third section of advanced options is the Additional modification to reads. The quality cutoff is used to trim bad quality bases from the reads before trimming adapter. Quality encoding tells the quality score encoding for the raw data. The Reads names prefix and suffix is used to add prefix and suffix to the read ID. Lastly, the Negative quality zero if checked will convert all negative quality score base to zero.
References
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17: 10-12.
...
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...