Page History
...
The 'cellranger count' pipeline from Cell Ranger v6.0.0[2] has been wrapped in Partek® Flow® as as Cell Ranger - Gene Expression task. It does not comprehensively cover all of the options and analysis cases Cell Ranger can handle for now, but converts FASTQ files from 'cellranger mkfastq' and performs alignment, filtering, barcode counting, UMI counting for single cell gene expression and Feature Barcode data. The output gene expression count matrix in .h5 format (both raw and filtered available for users to download in the output page of task details) then becomes the starting point for downstream analysis for scRNA-seq in Flow. For Feature Barcode data, Flow outputs a unified feature-barcode matrix that contains gene expression counts alongside Feature Barcode counts for each cell barcode.
...
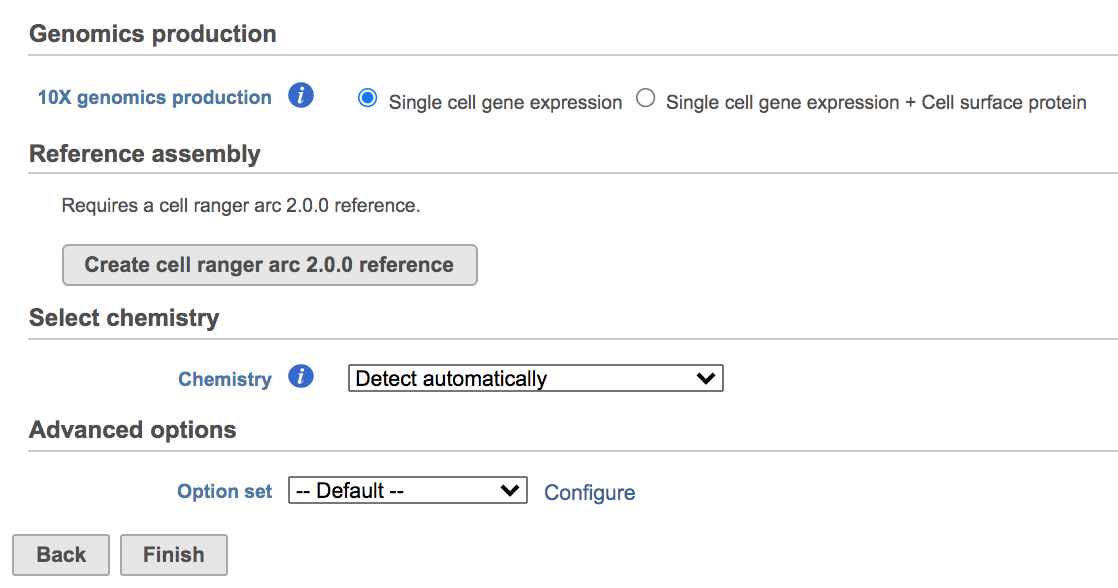
Once the Genomics production has been picked, users will be asked to create a Reference assembly if it is the first time to run the Cell Ranger - Gene Expression task (Figure 2). In Partek Flow, we will use Cell Ranger ARC 2.0.0 to create reference assembly for all 10x Genomics analysis pipelines. To create and use a reference assembly, Cell Ranger ARC requires a reference genome sequence (FASTA file) and gene annotations (GTF file), here is are the details.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...