Page History
...
This guide gives the basics of Partek® Flow® usage. Partek Flow can be installed in either a server, computer cluster or on the cloud. Regardless of where it's installed, it can be viewed using any web browser. We recommend using Google Chrome.
This guide covers:
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
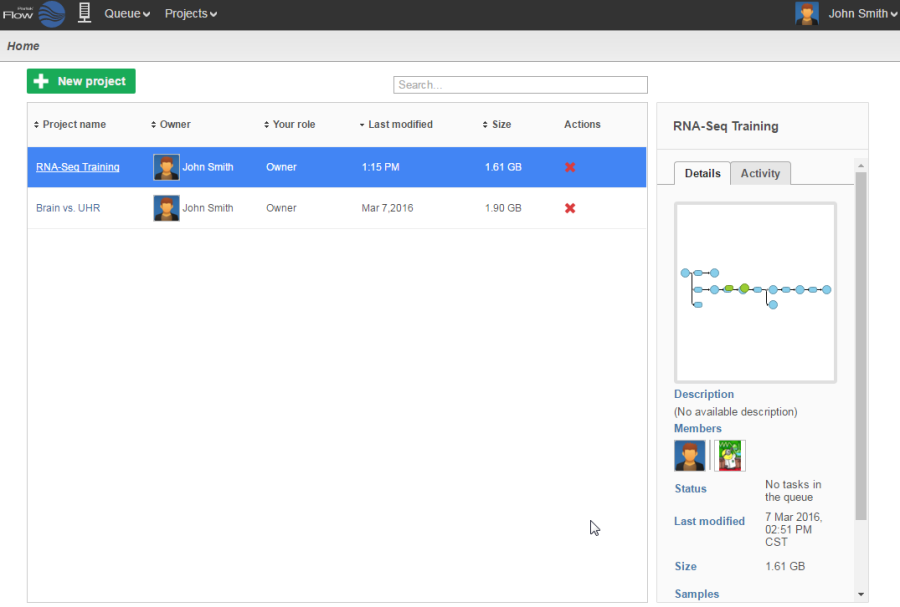
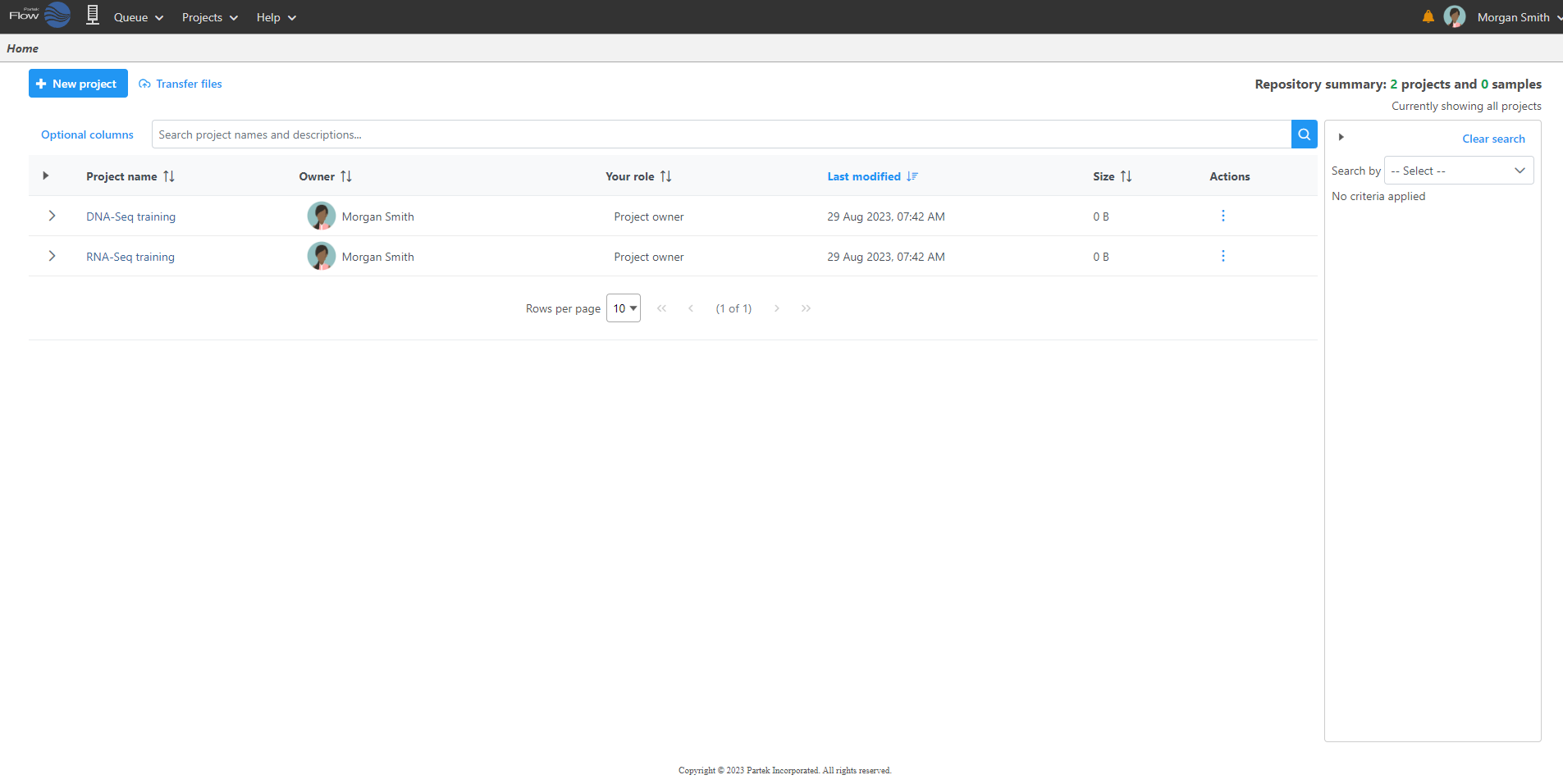
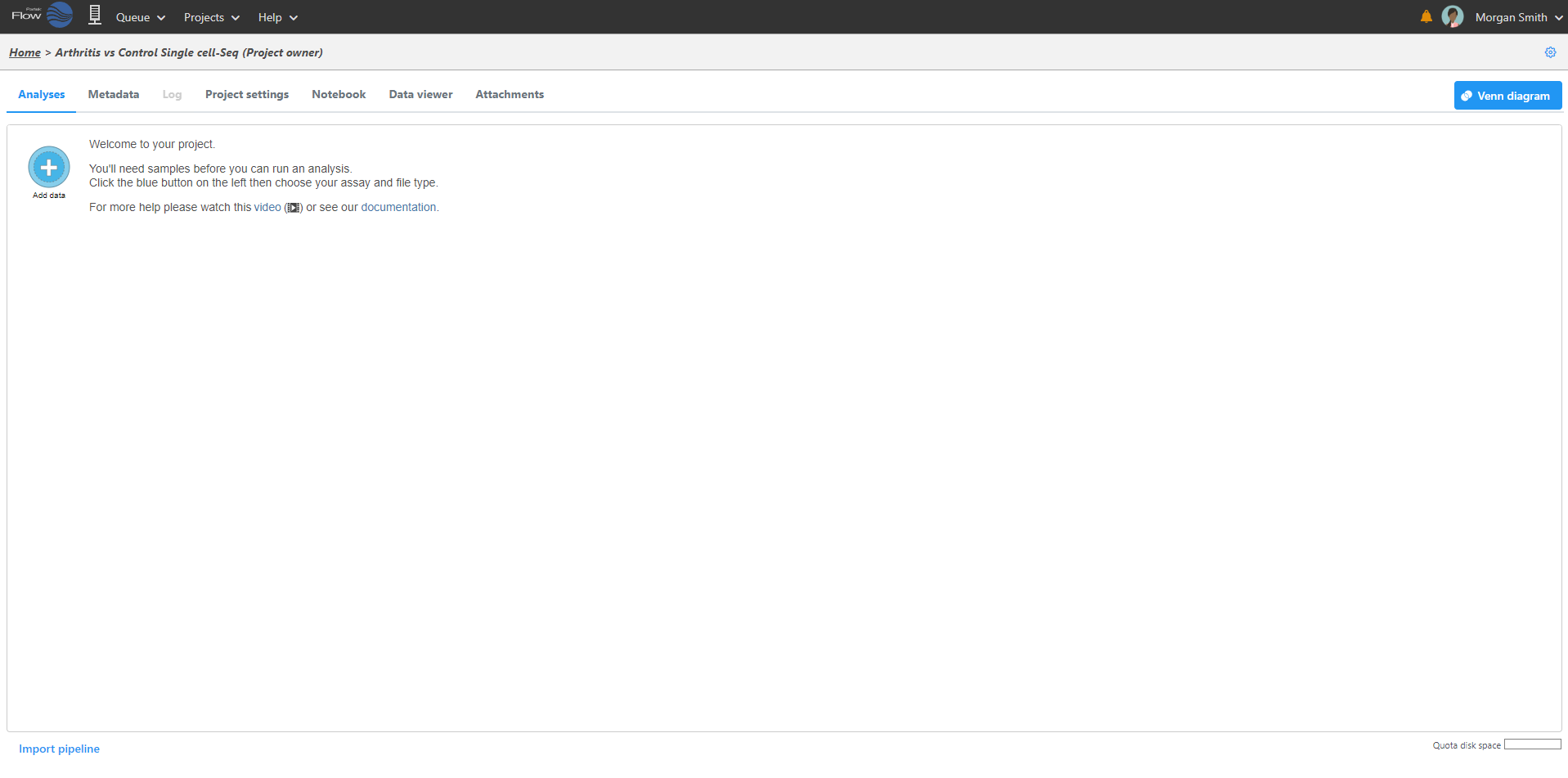
Logging in to your Partek Flow account will bring up the Home page (Figure 1). This page will show recent activities you've performed, recent projects you've worked on and pertinent details about each project.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Starting a new project
From the Home Page, click the New project button button . Assign Assign a name to the project and click the Create project button.
Uploading your dataset
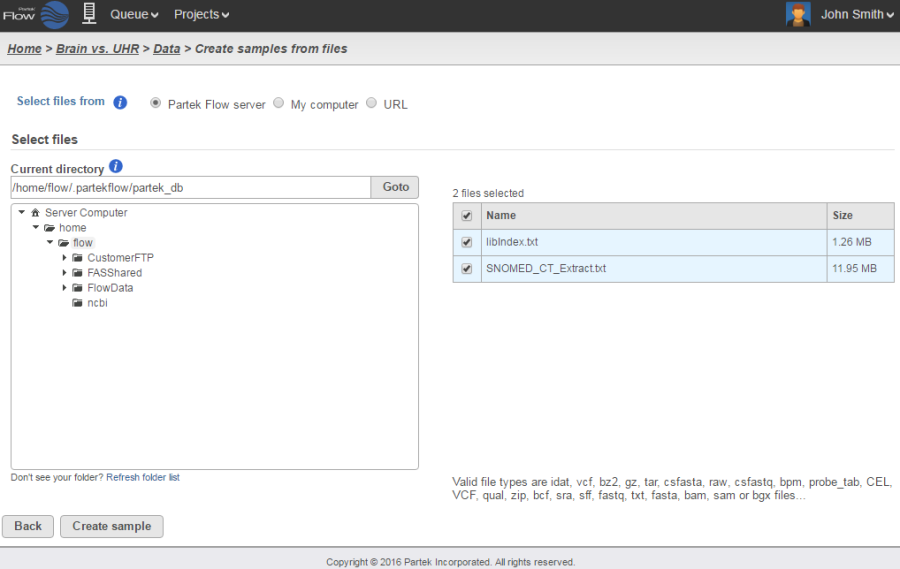
Upon creation of a new project, the Data tab Analyses tab will appear, prompting you to add samples to your project. Click the blue Add samplesdata button . Selecting Automatically create samples from files is the fastest way to create a sample (Figure 2).
Select the type of data (Single cell, Bulk, Other), choose the assay type, and select the data format. Partek Flow accepts various data types including unaligned and aligned Next Generation Sequencing (NGS) reads, and microarray data. . Use the Next button to proceed with import.
There are three ways you can upload the data (Figure 2):
- From your Partek Flow server Directly from My computer (i.e. the computer you are currently using to access Partek Flow) (500 MB limit)(click here for more information)
- From a URL
...
- From a GEO / ENA Bioproject (click here for more information)
| Numbered figure captions | ||||||
|---|---|---|---|---|---|---|
|
| |||||
Because genomics datasets are generally large, it is ideal to have the data copied in a folder directly accessible to the Partek Flow server. Make sure that the directory has the appropriate permissions for Partek Flow to read and write files in that folder. You may wish to seek assistance from your system administrator in uploading your data directly.
Select the files you would like to create samples from. Once they've been created, assign the corresponding sample attributes for each sample using the Metadata tab. The most efficient way to assign sample attributes is by clicking Assign sample attributes from a file and uploading a tab delimited text file. The file should contain a table with the following:
- The first row lists the attribute names (e.g. Treatment, Exposure) and
- The first column of the table lists the sample names (the sample names in the file must be identical to the ones listed in the Sample name column in the Data tab)
- List the corresponding attributes for each sample in the succeeding columns
Basic Partek Flow layout
The
...
Analyses Tab
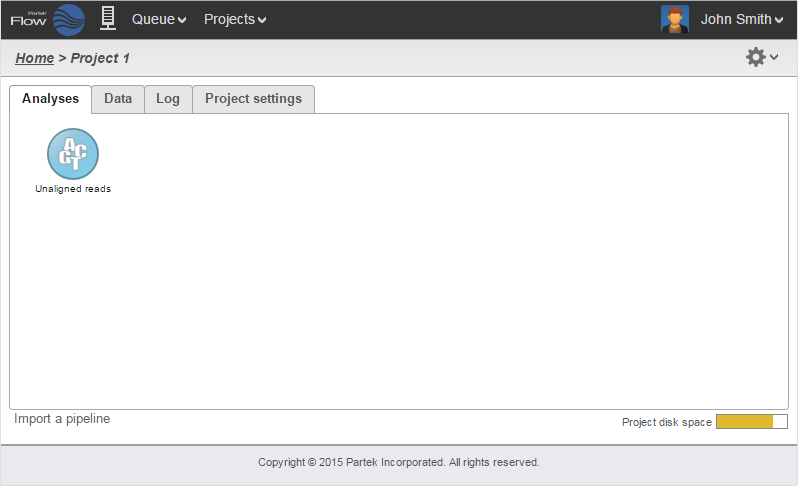
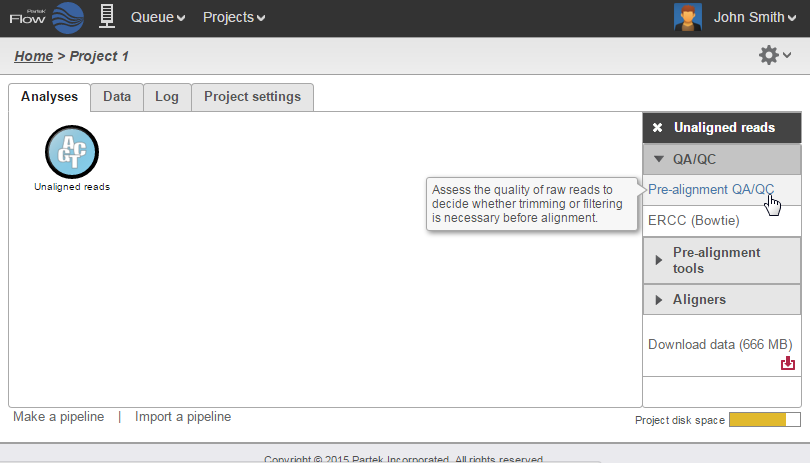
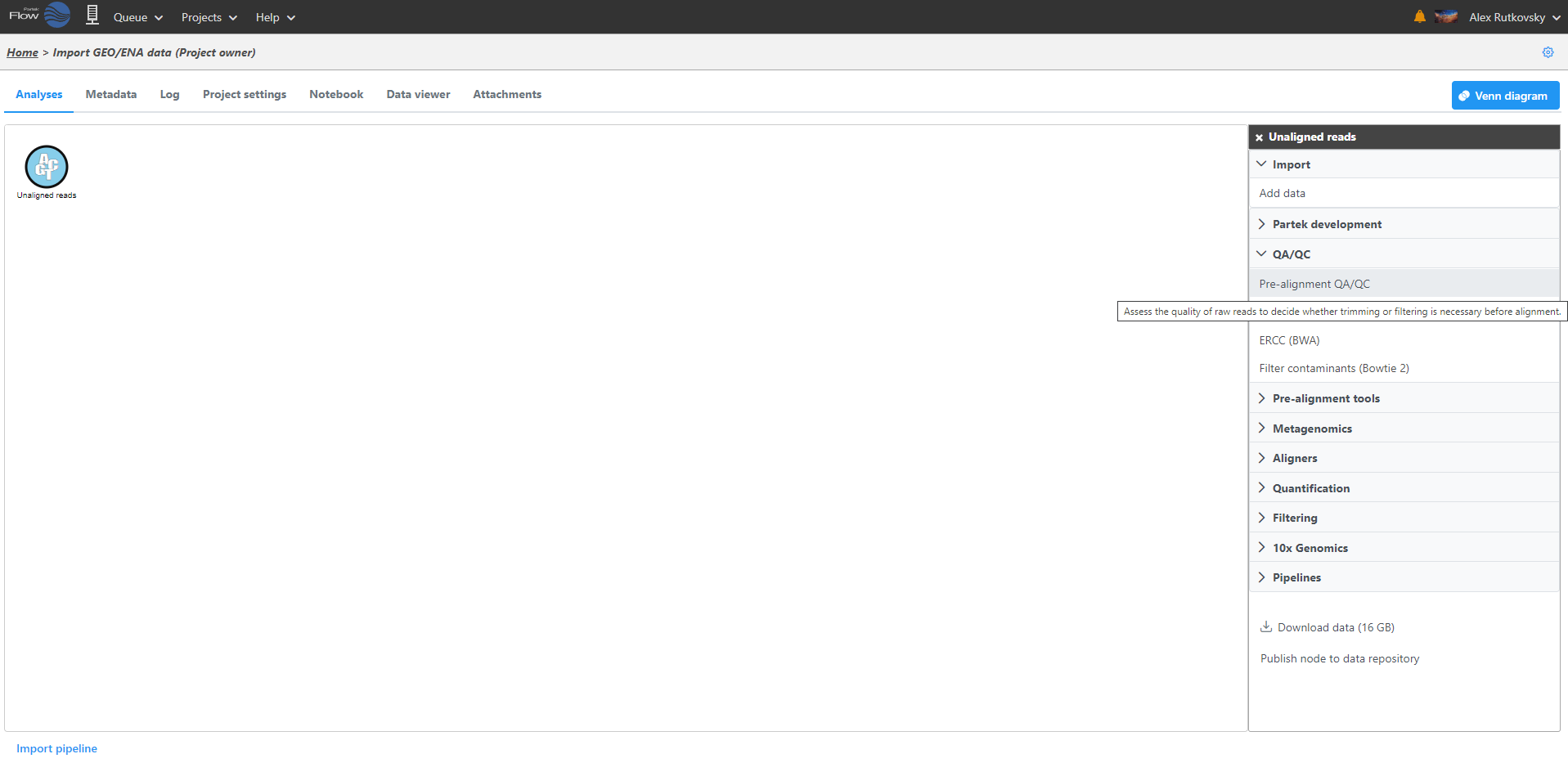
After samples have been added and associated with valid data files, a data node will appear in the Analysis Analyses tab (Figure 3). The Analysis Analyses tab is where you can invoke tasks, using the context sensitive menu on the right, and view the results of your analysis.
To add more data use the Add data task in the menu on the right or Add data in the Metadata tab. Once a task is performed, data can no longer be added to the project.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
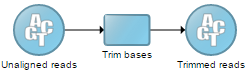
Data and task nodes
The Analyses tab contains two elements: data nodes (circles) and task nodes (rounded rectangles) connected by lines and arrows . Collectively, they represent a data analysis pipeline (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
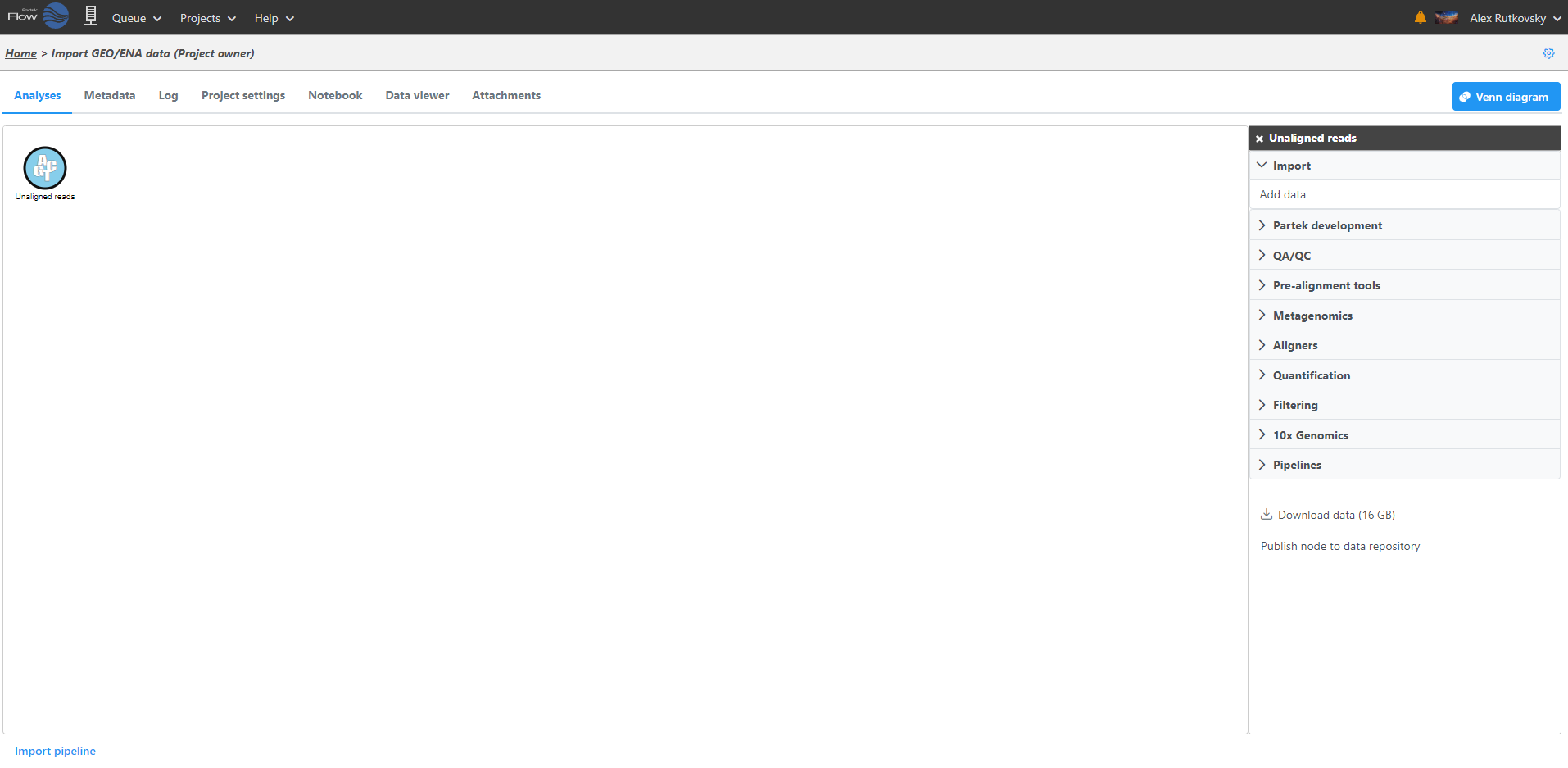
Clicking a data node brings up a context sensitive menu on the right (Figure 5). This menu changes depending on the type of data node. It will only present tasks which can be performed on that specific data type. Hover over the task to obtain additional information regarding each option.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
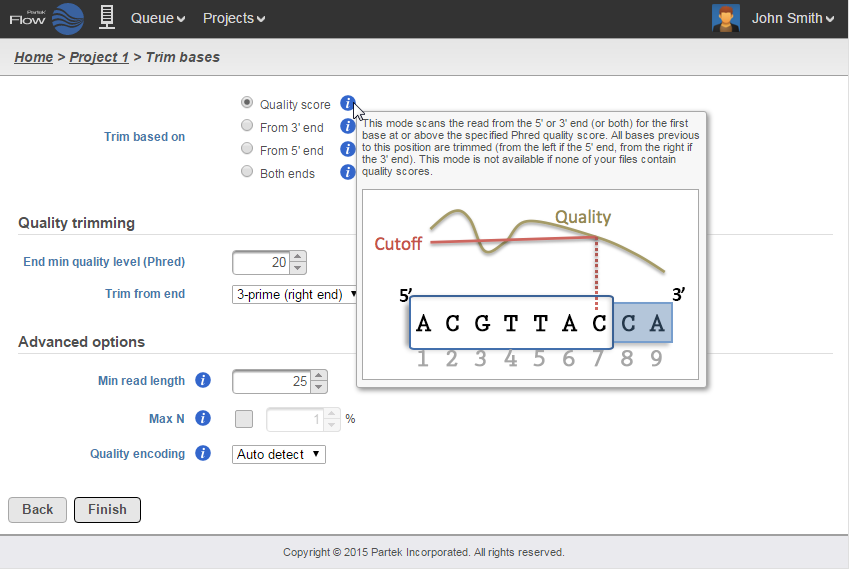
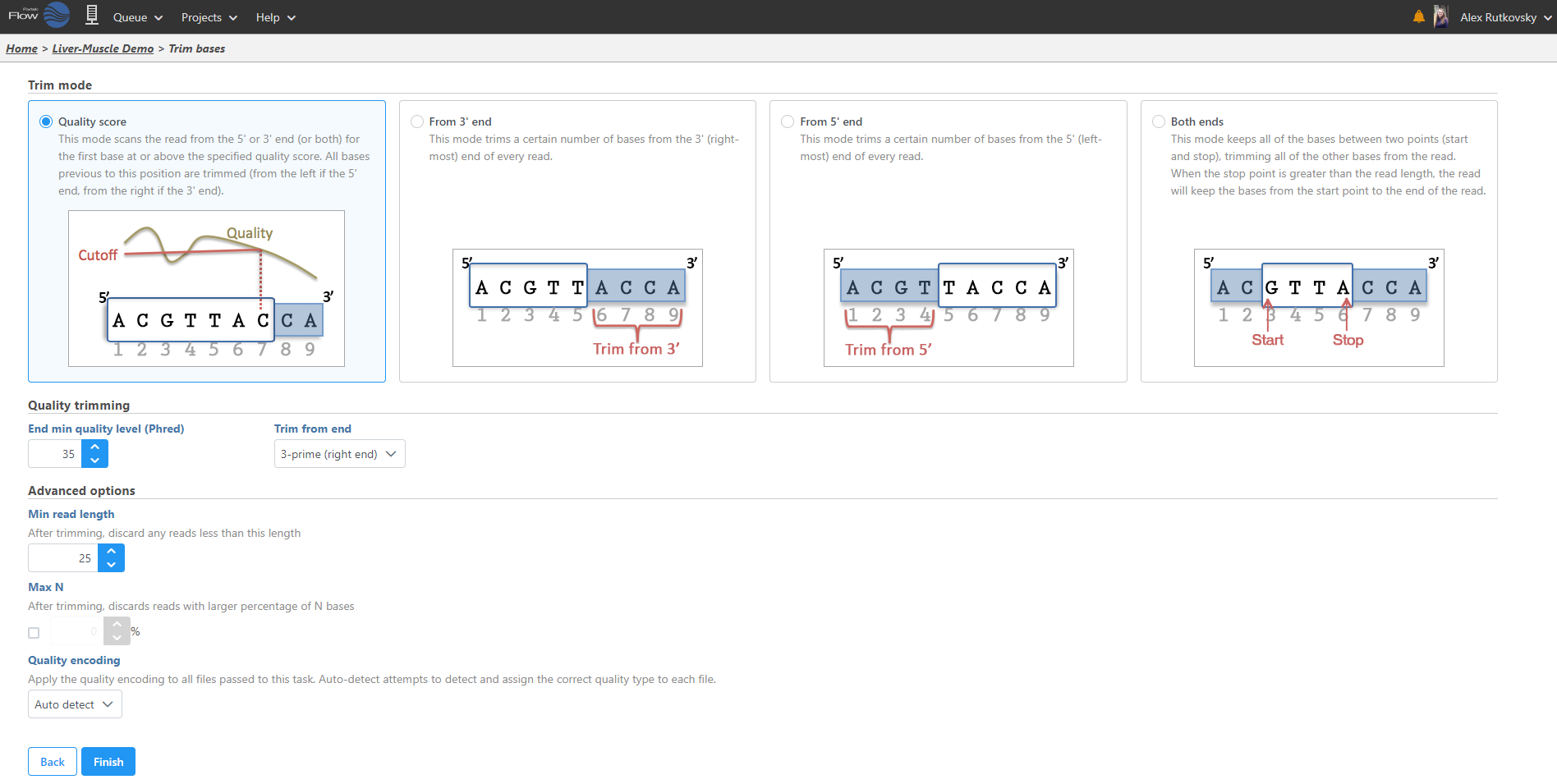
Select the task you wish to perform from the menu. When configuring task options, hover over the tab to obtain additional information regarding each option is available (Figure 6). When available, hover over Tooltips or click thevideo help for decision making. Click Finish to perform the task.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Depending on the task, a new data node may automatically be created and connected to the original data node. This contains the data resulting from the task. Tasks that do not produce new data types, such as Pre-alignment QA/QC, will not produce an additional data node.
To view the results of a task, click the task node and choose the Task report option on the menu.
Saving visualizations
Click the Click Save button on any visualization to export a publication-quality image (Figure 7).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Downloading your data
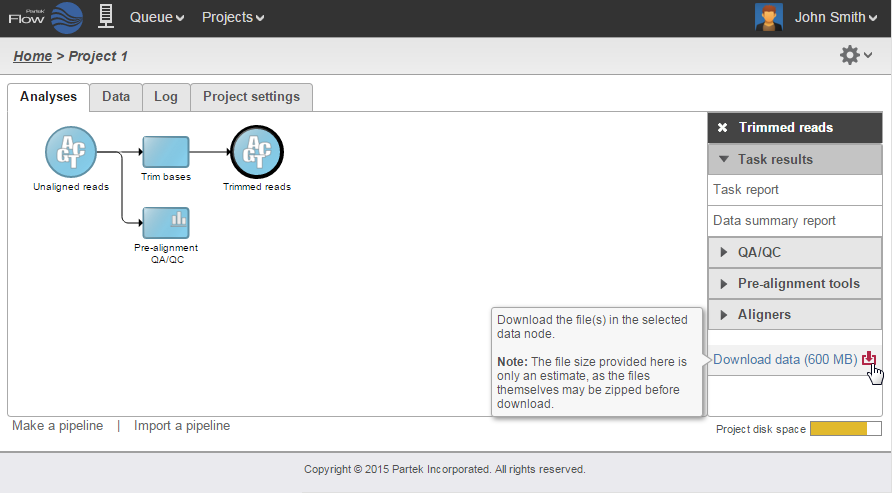
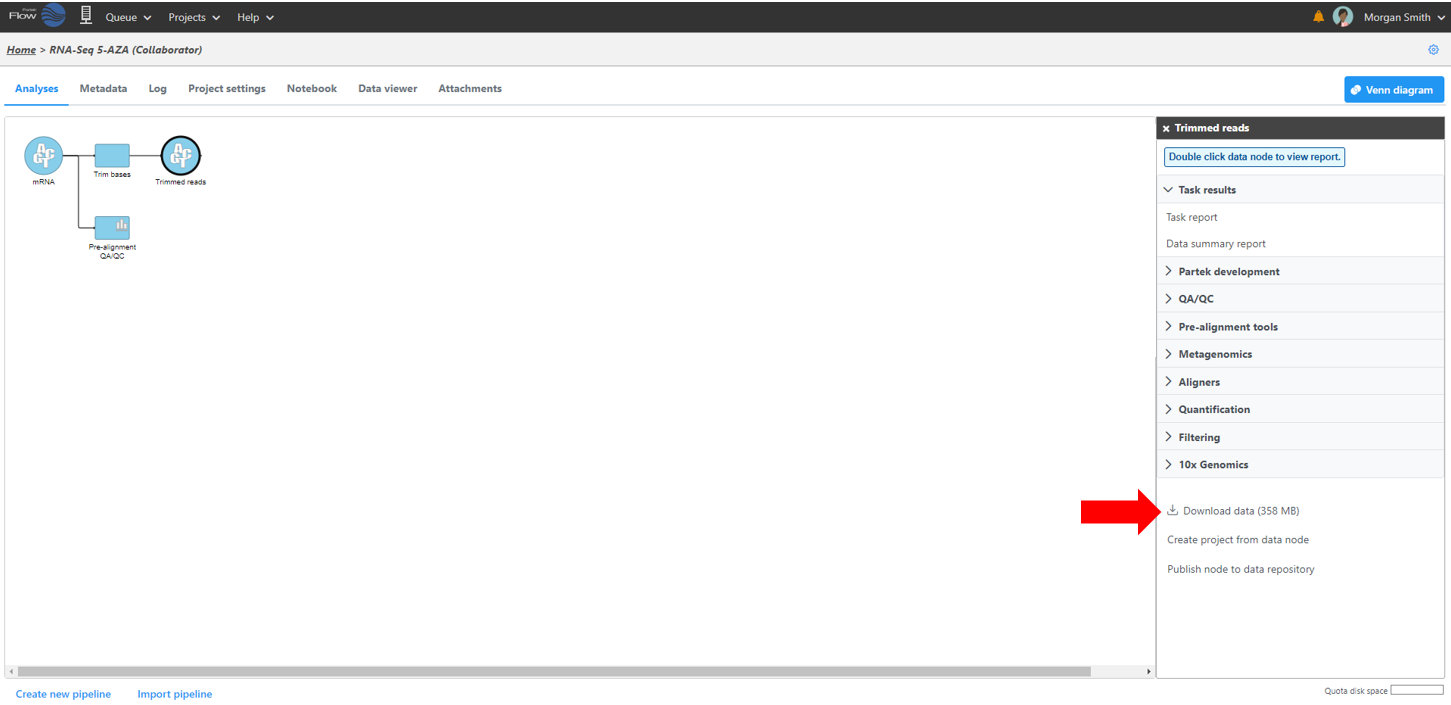
Data associated with any data node can be downloaded by clicking the node and choosing Download data at the bottom of the task menu (Figure 8). Compressed files will be downloaded to the local computer where the user is accessing the Partek Flow server. Note that bigger files (such as unaligned reads) would take longer to download. For guidance, a file size estimate is provided for each data node. Downloaded files can be seamlessly imported in Partek® Genomics Suite®.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Partek Flow in action
Watch a webinar of how to set-up a simple RNA-Seq project.
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|