...
| Numbered figure captions |
|---|
| SubtitleText | Illumina BeadArray Methylation workflow |
|---|
| AnchorName | Illumina BeadArray Methylation workflow |
|---|
|
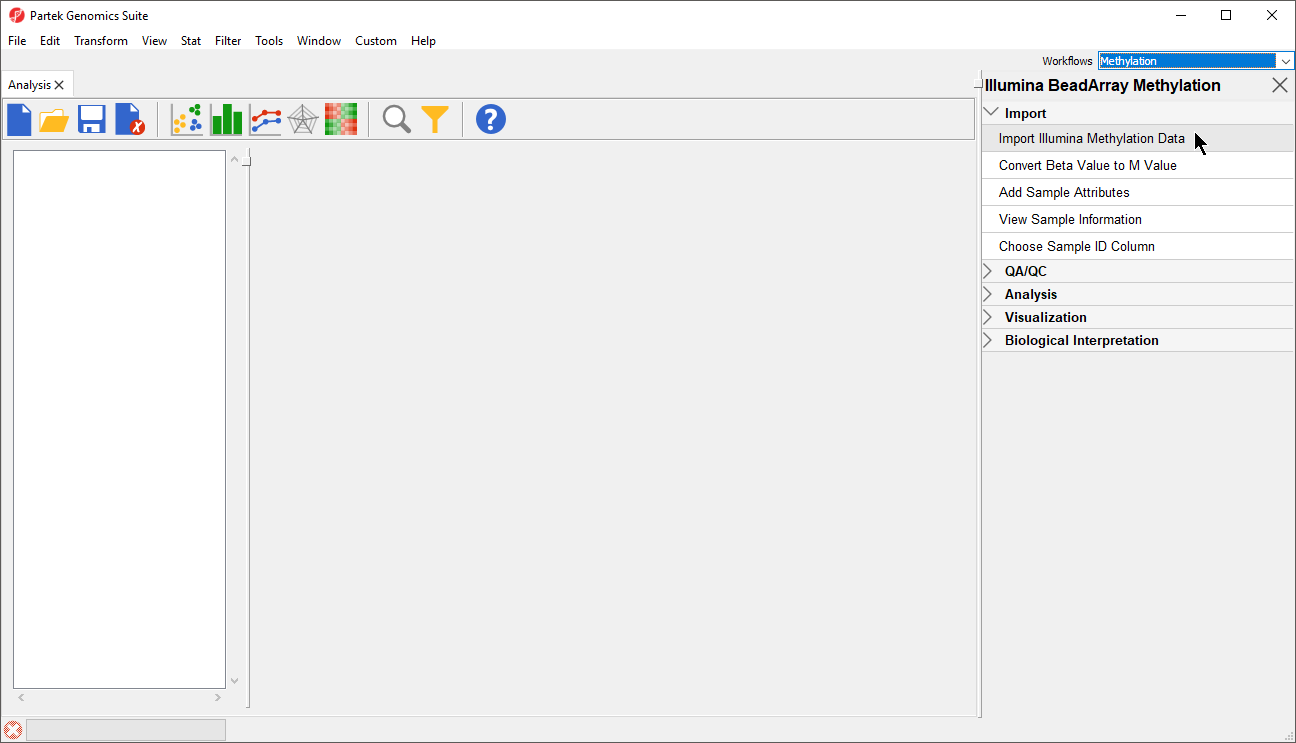 Image Removed Image Removed Image Added Image Added
|
- Select Import Illumina Methylation Data to bring up the Load Methylation Data dialog
...
| Numbered figure captions |
|---|
| SubtitleText | Selecting .idat files to import |
|---|
| AnchorName | Import Illumina iDAT data |
|---|
|
 Image Removed Image Removed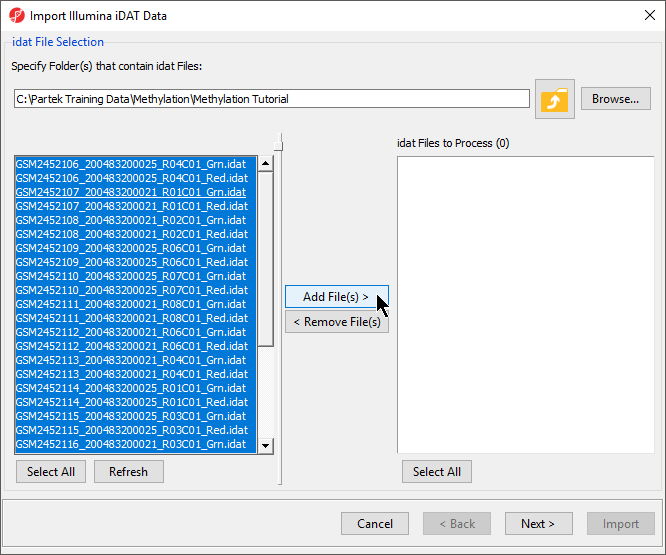 Image Added Image Added
|
- Select Add File(s) > to move the files to the idat Files to Process pane of the Import Illumina iDAT Data dialog (Figure 6)
| Numbered figure captions |
|---|
| SubtitleText | Confirming selection of .idat files for import |
|---|
| AnchorName | Selected .idat files for import |
|---|
|
 Image Removed Image Removed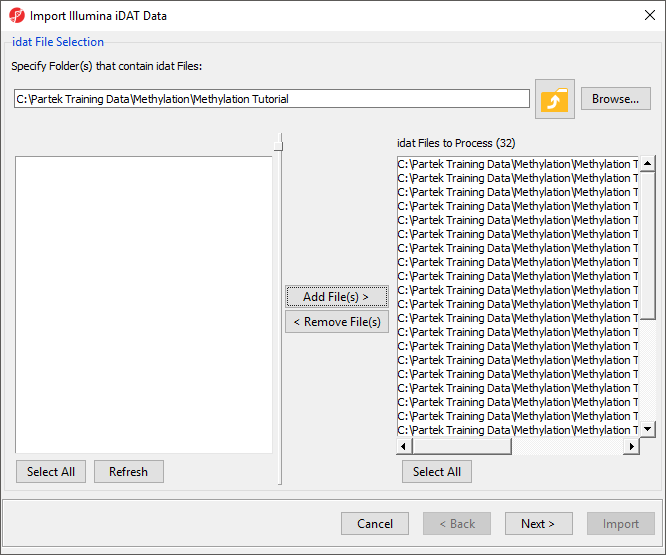 Image Added Image Added
|
The following dialog (Figure 7) deals with the manifest file, i.e. probe annotation file. If a manifest file is not present locally, it will be downloaded in the Microarray libraries directory automatically. The download will take place in the background, with no particular message on the screen and it may take a few minutes, depending on the internet connection. In the future, you may want to reanalyze a data set using the same version of the manifest file used during the initial analysis, rather than downloading an up-to-date version. To facilitate this, the Manual specify option in the Manifest File section allows you to specify a specific version. For this tutorial, we will leave this on the default settings.
| Numbered figure captions |
|---|
| SubtitleText | Selecting manifest file and output file |
|---|
| AnchorName | Import Illumina iDAT Data dialog |
|---|
|
 Image Removed Image Removed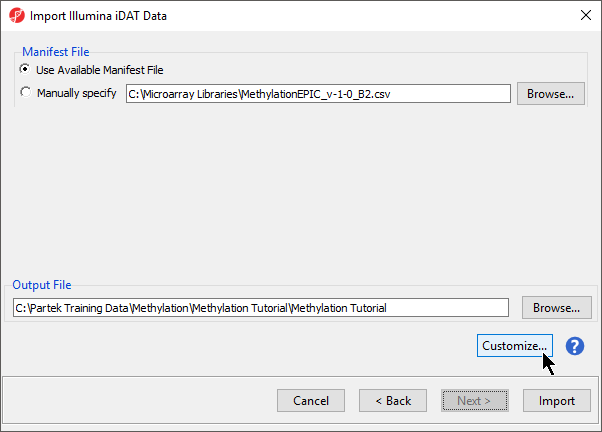 Image Added Image Added
|
By default the output file destination is set to the file containing your .idat files and the name matches the file folder name. The name and location of the output file can be changed using the Output File panel.
...
In the Algorithm tab of the Advanced Import Options dialog (Figure 8), there are two filtering options and five normalization options available. Select the ( Image Removed) next to each method for details. The filters allow you to exclude probes from the X and Y chromosomes or based on detection p-value. In this tutorial, we have male and female samples, so we will apply the X and Y chromosome Filter. We will also filter probes based on detection p-value to exclude low-quality probes.
Image Removed) next to each method for details. The filters allow you to exclude probes from the X and Y chromosomes or based on detection p-value. In this tutorial, we have male and female samples, so we will apply the X and Y chromosome Filter. We will also filter probes based on detection p-value to exclude low-quality probes.
- Select Exclude X and Y Chromosomes
- Select Exclude probes using detection p-value and leave the default settings of 0.05 and 1 out of 16 samples.
We recommend using the default optionsoption for normalization; however, advanced users can select their preferred normalization method. Select the ( Image Added) next to each normalization option for details. If you want to import probe intensity, raw probe intensity, probe signals, raw probe signals, or anti-log probe intensity values, they can be added to the data import using the Outputs tab of the Advanced Import Options dialog. Probe intensities and raw probe intensities can be used for advanced troubleshooting purposes and antilog probe intensities can be used for copy number detection. The Outputs tab of the Advanced Import Options dialog also has an option to create NCBI GEO submission spreadsheets from your imported data. For this tutorial, we do not need to import any of these values or create GEO submission spreadsheets.
Image Added) next to each normalization option for details. If you want to import probe intensity, raw probe intensity, probe signals, raw probe signals, or anti-log probe intensity values, they can be added to the data import using the Outputs tab of the Advanced Import Options dialog. Probe intensities and raw probe intensities can be used for advanced troubleshooting purposes and antilog probe intensities can be used for copy number detection. The Outputs tab of the Advanced Import Options dialog also has an option to create NCBI GEO submission spreadsheets from your imported data. For this tutorial, we do not need to import any of these values or create GEO submission spreadsheets.
| Numbered figure captions |
|---|
| SubtitleText | Advanced Import Options offers choice of normalization method and additional data outputs |
|---|
| AnchorName | Advanced Import Options iDAT |
|---|
|
 Image Removed Image Removed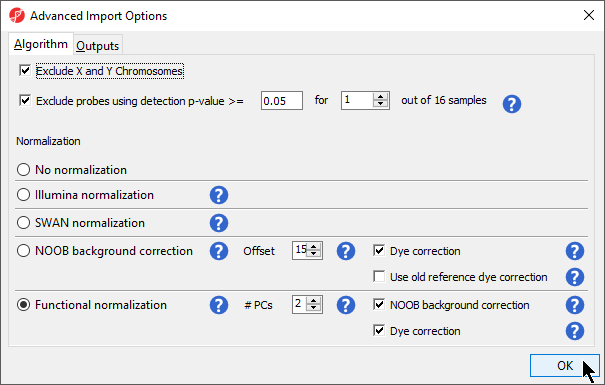 Image Added Image Added
|
- Select OK to close the Advanced Import Options dialog dialog
- Select Import on the Import Illumina iDAT data dialog
The imported and normalized data will appear as a spreadsheet 1 (Methylation Tutorial) (Figure 9)
| Numbered figure captions |
|---|
| SubtitleText | Viewing the imported methylation data in a spreadsheet |
|---|
| AnchorName | Imported .idat File Spreadsheet |
|---|
|
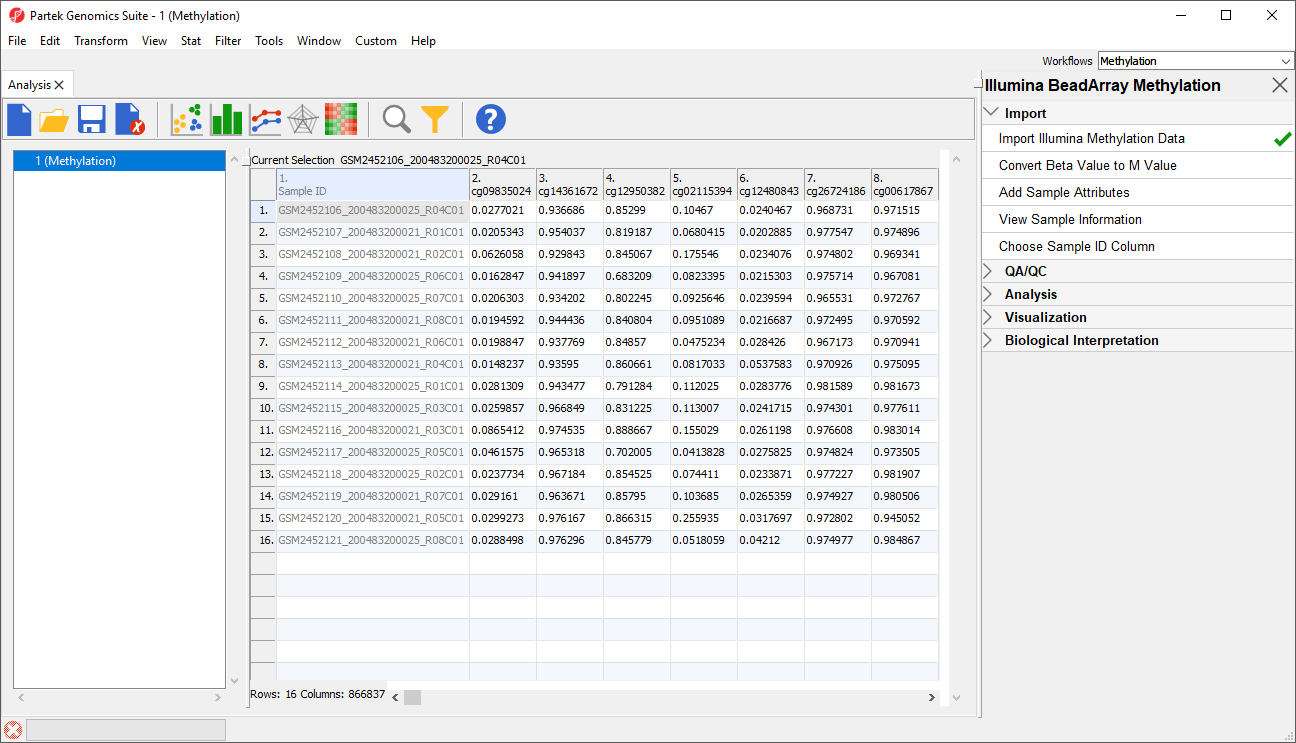 Image Removed Image Removed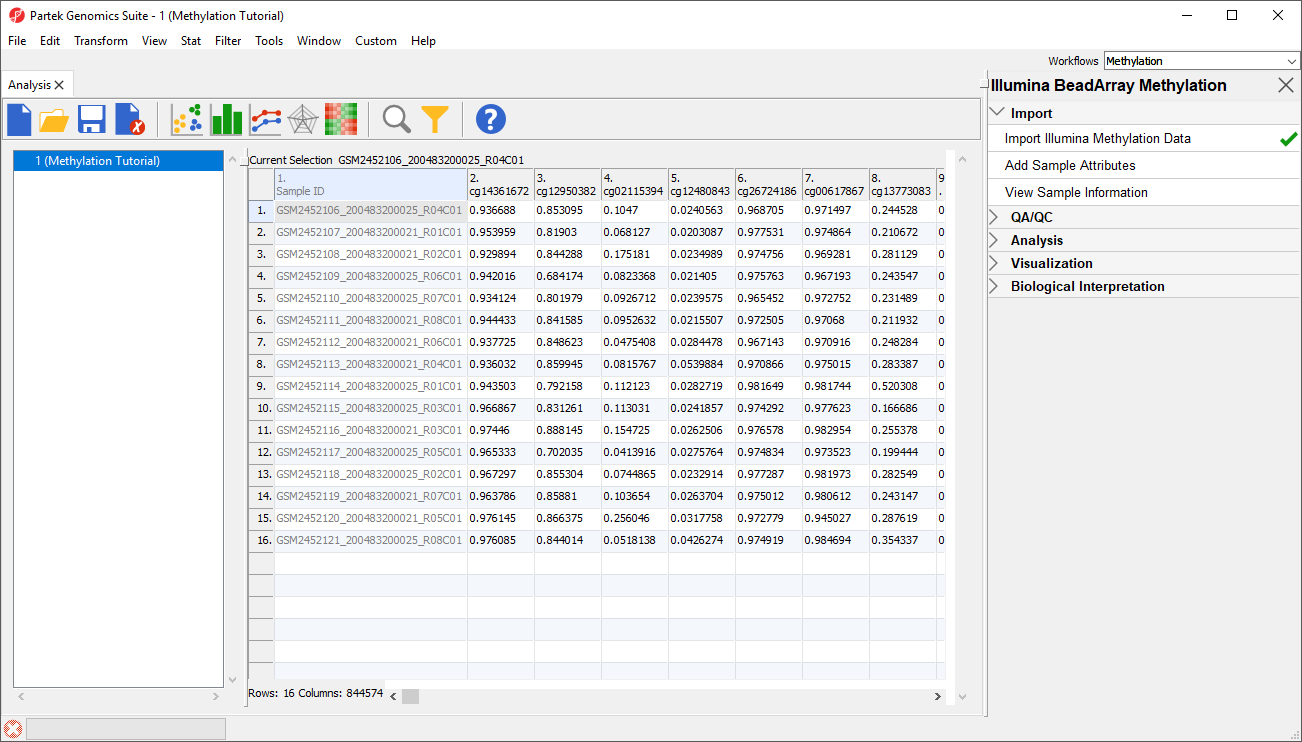 Image Added Image Added
|
...











