Page History
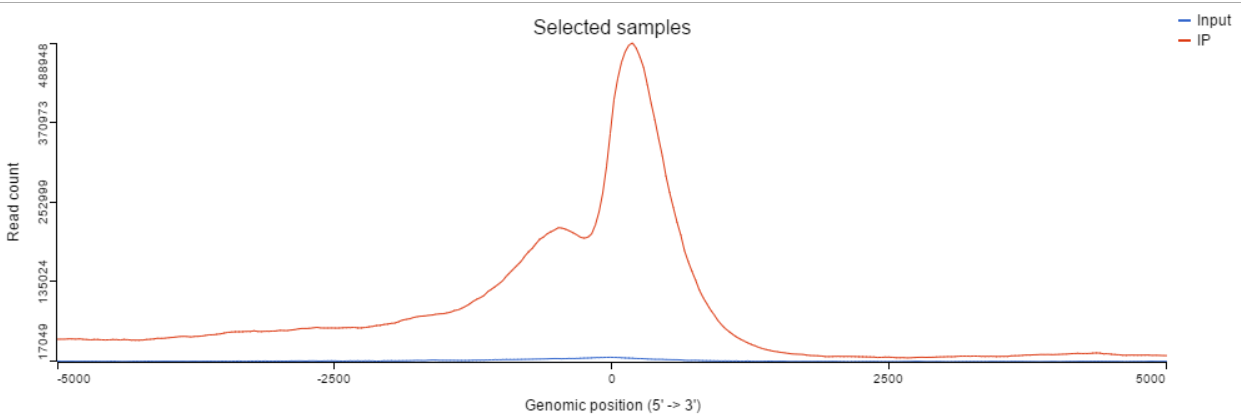
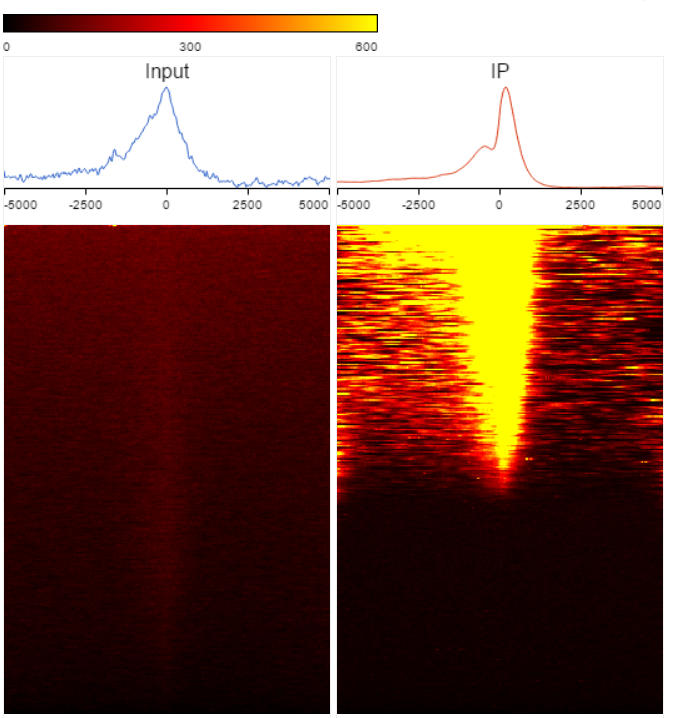
TSS plot is available on annotated peak data node. After the peaks are annotated with transcripts, coverage of the reads across the genes overlapped with the regions will be calculated, this information is presented by profile plot (Figure 1) and heatmap (Figure 4).
| Table of Contents | ||||||
|---|---|---|---|---|---|---|
|
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
TSS profile plot
In the profile plot, the Y-axis is the total read counts, X-axis is the window defined in the annotation task, mid point is the transcription start site (TSS) of all the genes overlapped with the peak regions, the start and end point of X-axis is promoter upstream/downstream limit specified in the annotate peaks task (Figure 2). Each line represents a selected sample total read counts at each position, all the selected samples are shown in the plot to compare.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
TSS heatmap
Heatmap is another way to view the coverage of gene body overlapped with the peaks (Figure 4). The X-axis represents the same profile plot, actually each samples profile plot is display again on the top of the heatmap, the max on the Y-axis are different in different samples. In the heatmap, each row represents a gene, color represents the total read count at each position, genes are sorted based on total read counts in descending order from the top to the bottom.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...