Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Select the 10x assay type and choose CytAssist gene expression if you are using the Visium CytAssist gene expression library.
For both Space Ranger or Cell Ranger tasks, a Reference assembly is required (Figure 2). To define the Reference assembly, first select the Genome build for the organism of interest, then select the annotation Index. In this manner, custom libraries can be created (e.g. keep the Genome build but change the annotation Index).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
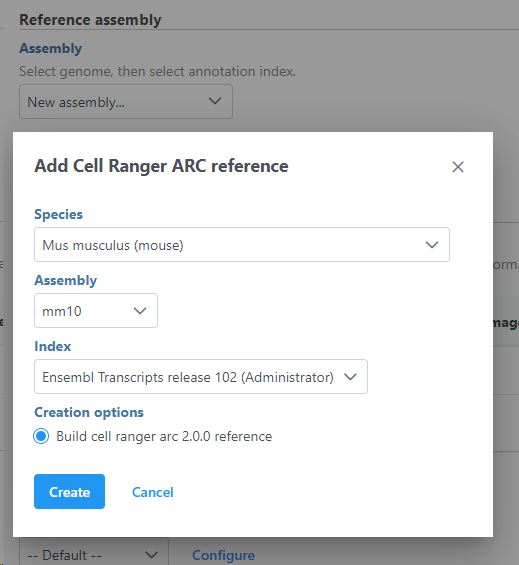
To add a new species genome, choose New assembly from the drop down for Genome build which will open a new window with configuration options to edit, then click Create (Figure 3).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The sample table under Input options has one row per sample (Figure 2). Image file is required, and that is a single hematoxylin and eosin brightfield image in either .jpg or .tiff format. Click on the Browse button under Browse image file and the file browser will come up. Point to the image file and push Continue. Formalin-fixed paraffin-embedded (FFPE) image files require the Probe set file otherwise it is optional; it is a .csv file specifying the probe set used (=target panel).
...