Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
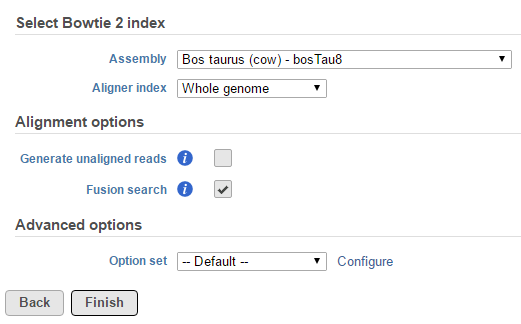
The output is dumped to the Fusion results (Figure 2), which is a part of TopHat 2 results (in addition to Aligned reads node and, optionally, Unaligned reads node).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
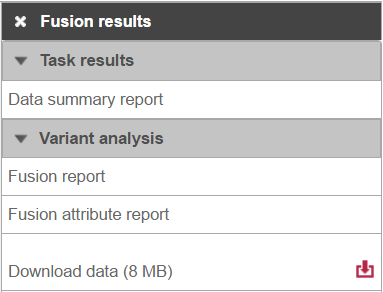
Selecting the Fusion results node opens the toolbox, with four options (Figure 3): Data summary report, Fusion report, Fusion attribute report, and Download data.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
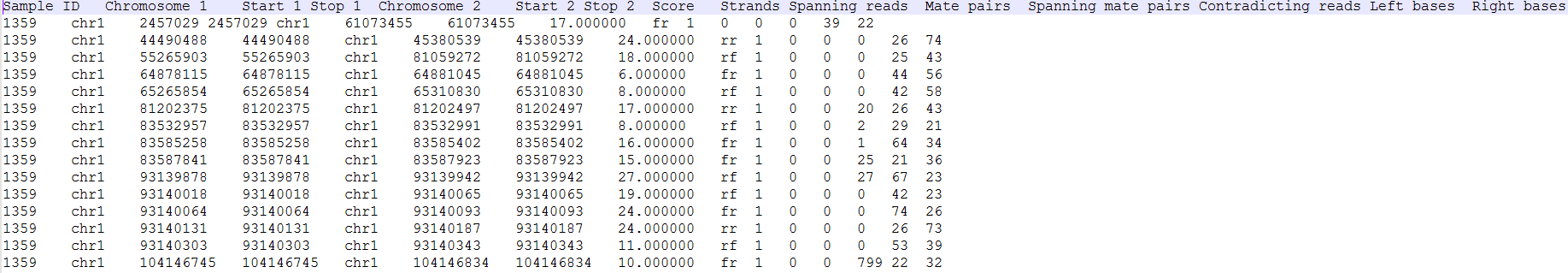
Clicking on the Download data results in download of a .fusion file to the local computer. The file is human readable and can be opened in a text editor (example in Figure 4). For details refer to TopHat-Fusion documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
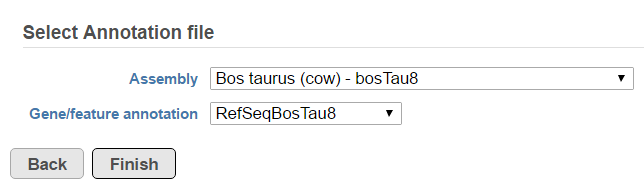
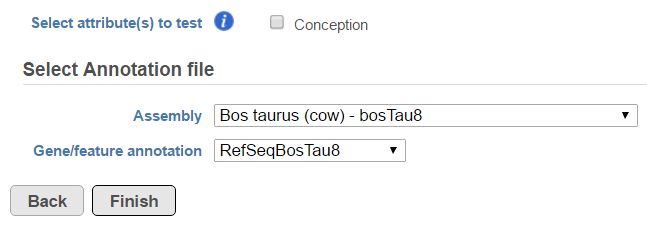
Fusion report provides an annotated report on detected fusion genes. For that purpose an annotation file needs to be specified first (Figure 5).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The result of annotation is the Fusion report task node as seen in Figure 6.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
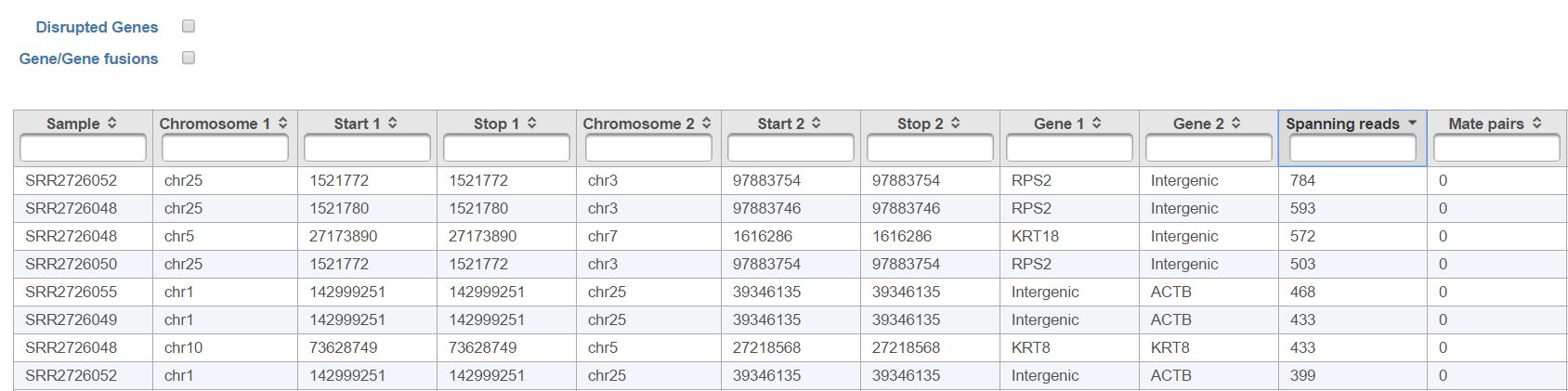
The list of annotated fusion genes, in a form of Fusion report (Figure 7), can be obtained by first selecting the Fusion report task node and then the Task report link from the toolbox. Each row of the table in Figure 7 is a potential fusion event, with the columns providing the following information.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Checkboxes Disrupted Genes and Gene/Gene fusions are filter tools. When selected Disrupted Genes removes all the rows (fusion events) which have no gene assigned to it, i.e. which merge two intergenic regions. However, if there is a fusion between a gene and an intergenic region, it will be kept in the table. Gene/Gene fusions filters in only those fusion events which have an annotated gene on both sided of the breakpoint. In the other words, only gene to gene fusions are kept in the table.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new data node, Fusion attribute report, is generated in the Analysis tab (Figure 9) and it provides access to the Task report link in the toolbox.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
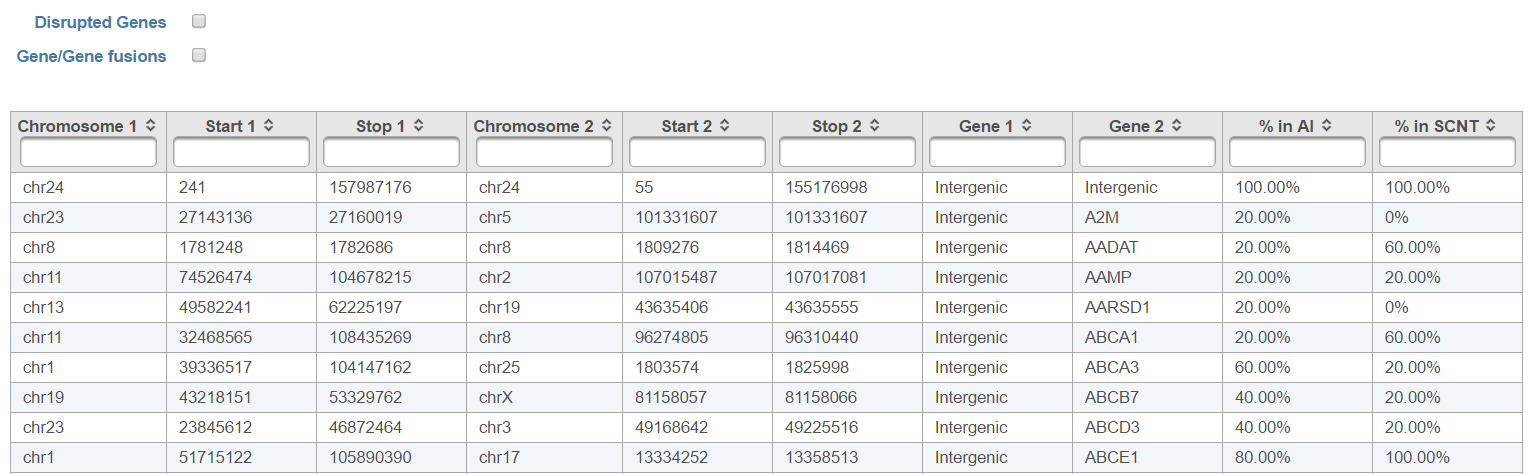
The output, Fusion report table (Figure 10) resembles the basic TopHat-Fusion output (Figure 7); each row of the table is a single fusion event while the information on the merged segments is on the columns.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
STAR Algorithm
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
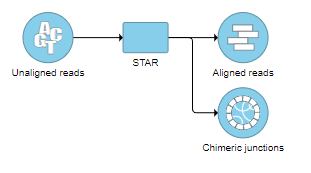
The output is associated with the Chimeric results data node (Figure 12), which is a part of STAR results (in addition to Aligned reads node and, optionally, Unaligned reads node).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
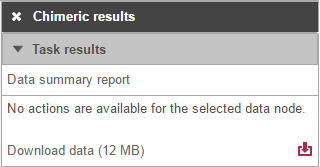
Selecting the Chimeric results node opens the toolbox (Figure 13) with twooptions: Data summary report or Download data.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
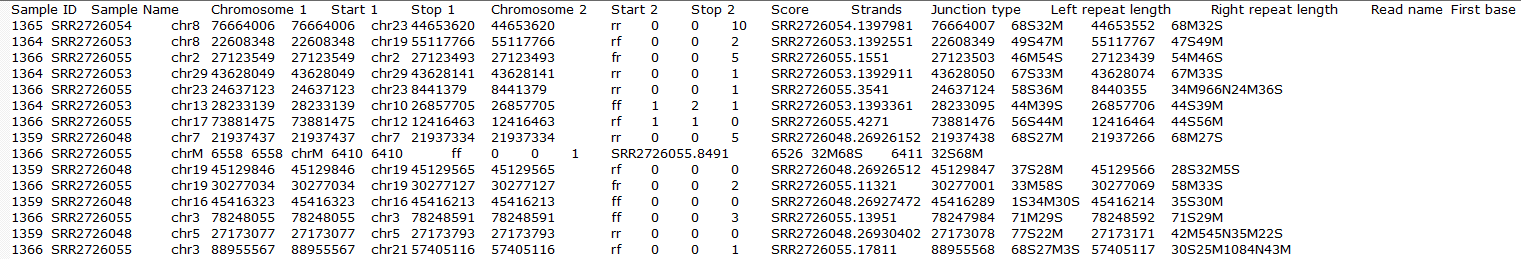
Clicking on the Download data results in download of a .fusion file to the local computer. The file is human readible and can be opened in a text editor (example in Figure 14). For details refer to STAR's documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
References
- Annala MJ, Parker BC, Zhang W, Nykter M. Fusion genes and their discovery using high throughput sequencing. Cancer Lett. 2013;340:192-200.
- Costa V, Aprile M, Esposito R, Ciccodicola A. RNA-Seq and human complex diseases: recent accomplishments and future perspectives. Eur J Hum Genet. 2013;21:134-142.
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biology. 2011;12:R72
- TopHat-Fusion. An algorithm for discovery of novel fusion transcripts. http:// http://tophat.cbcb.umd.edu/fusion_index.html Accessed on April 25, 2014
- Dobin A, Davies CA, Schlesinger F et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15-21.
...