Join us for an event September 26!
How to Streamline RNA-Seq analysis and increase productivity—point, click, and done
Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
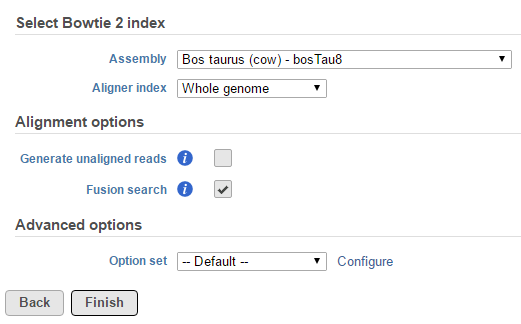
The output is dumped to the Fusion results (Figure 2), which is a part of TopHat 2 results (in addition to Aligned reads node and, optionally, Unaligned reads node).
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
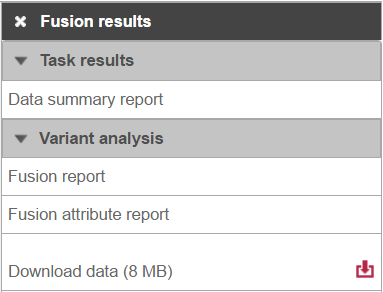
Selecting the Fusion results node opens the toolbox, with Variant detection options four options (Figure 8)3): Data summary report, Fusion report, Fusion attribute report, and Download data.
| Numbered figure captions | |
|---|---|
|
...
|
...
...
| |
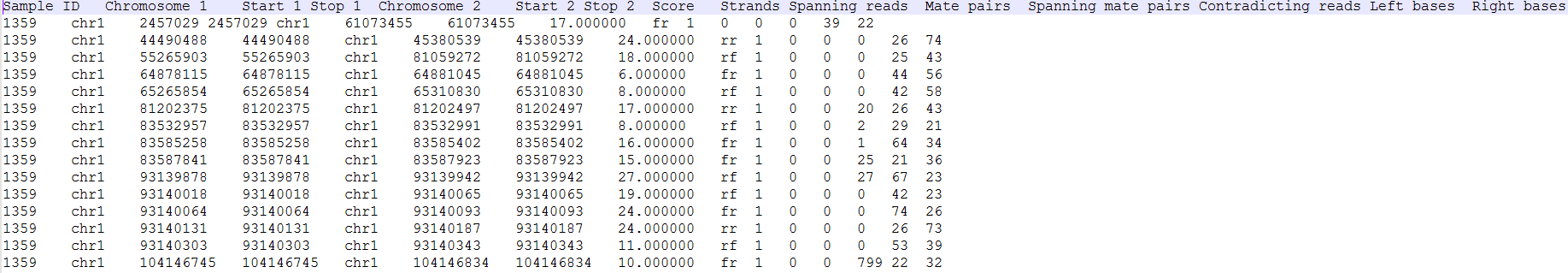
Clicking on the Download data results in download of a .fusion file to the local computer. The file is human readable and can be opened in a text editor (example in Figure 4). For details refer to TopHat-Fusion documentation.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
|
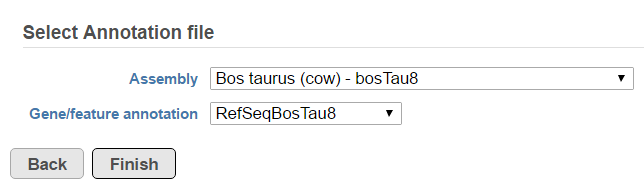
Fusion report provides an annotated report on detected fusion genes. For that purpose an annotation file needs to be specified first (Figure 95).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
The result of annotation is the Fusion report task node as seen in Figure 106.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
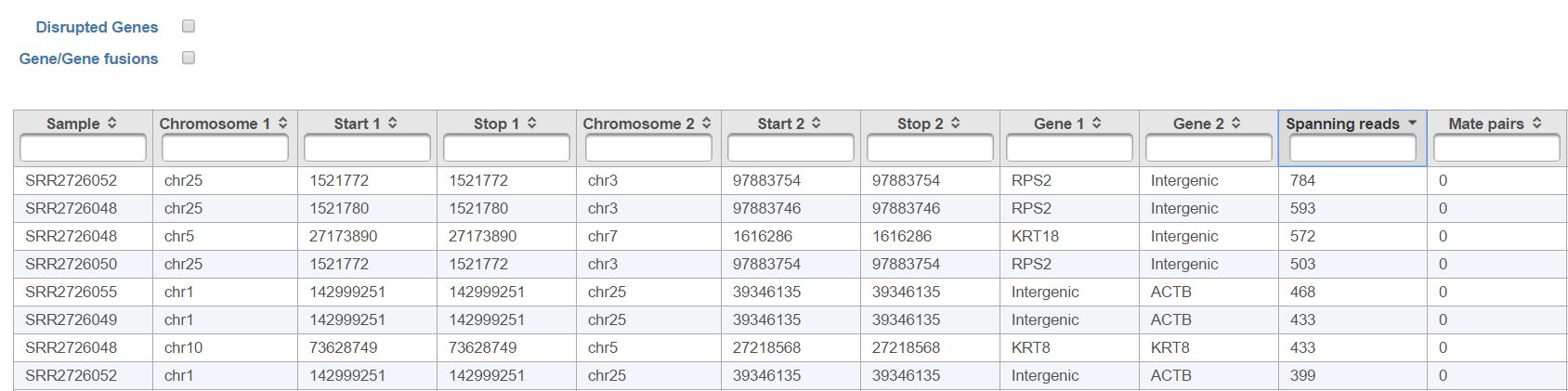
The list of annotated fusion genes, in a form of Fusion report (Figure 11), can be obtained by first selecting the Fusion report task node and then the Task report link from the toolbox. Each row of the table in Figure 11 7 is a potential fusion event, with the columns providing the following information.
- Sample ID: sample in which the fusion event was identified;
- Chromosome 1: chromosome hosting the left part of the fusion;
- Start 1: end of the first segment
- Stop 1:
- Chromosome 2: chromosome hosting the right part of the fusion;
- Start 2:
- Stop 2:
- Score: fusion score as defined in the original TopHat-Fusion report (3);
- Type1: genomic section of the left-hand part of the fusion;
- Gene1: gene on the left side of the fusion;
- Transcript1: affected transcript of the Gene1;
- Type2: genomic section of the right-hand part of the fusion;
- Gene2: gene on the right side of the fusion;
- Transcript2: affected transcript of the Gene2
- Loci: coordinates of the fusion event (a dash indicates genes on different chromosomes, while a colon indicates that both genes are on the same chromosome with the distance between the parts being given after the colon);
- Strands: orientation of the two chromosomes (e.g. ff indicates that both chromosomes are in forwarding direction);
- Spanning reads: the number of reads spanning the fusion.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Moreover, Fusion attribute report, when invoked from the Fusion results node, displays a report on attributes of detected fusion genes. Attributes to be tested for association with the fusion should be specified first (Figure 12).
...