Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
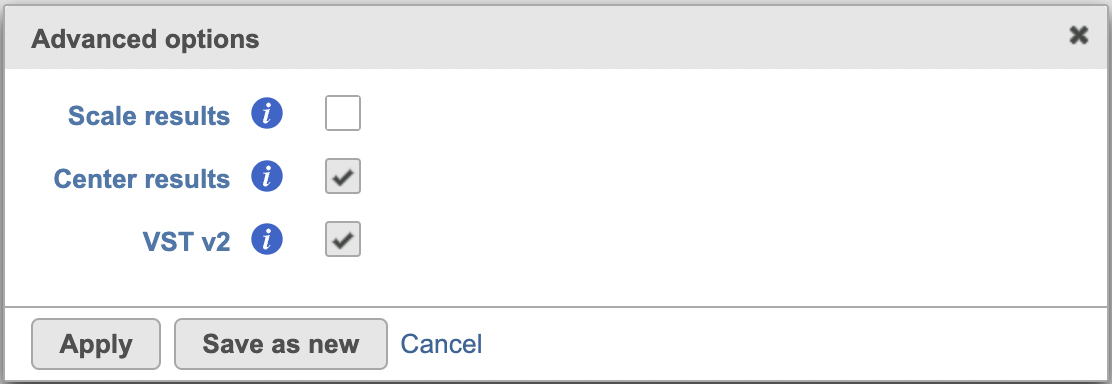
Scale results: Whether to scale residuals to have unit variance; default is FALSE
Center results: When set to Yes, center all the transformed features to have zero mean expression. Default is TRUE.
VST v2: Default is TRUE. When set to 'v2', it sets method = glmGamPoi_offset, n_cells=2000, and exclude_poisson = TRUE which causes the model to learn theta and intercept only besides excluding poisson genes from learning and regularization; If default is unchecked, it uses the original sctransform model (v1), it will only generate SC scaled data node.
There are two data nodes generated from this task (if VST v2 option is checked as default):
SC scaled data: it is a matrix of normalized values (residuals) that by default has the same size as the input data set. This data node that are is used to perform downstream exploratory analysis e.g. PCA, Seurat3 integration etc (Figure 3), this data node is not recommend to use for differential analysis.
SC corrected data: is equivalent to the ‘corrected counts’ in data slot generated after PrepSCTFindMarkers task in the SCT assay in Seurat object. It is used for downstream differential expression(DE) analyses (Figure 3).
...