...
Obtain and add files to the project
The project includes Human Breast Cancer (Block A Section In Situ Replicate 1) and Human Breast Cancer (Block A Section In Situ Replicate 2) output files files in one project.
- Obtain the filtered Count matrix files (h5 or HDF5) files and Spatial outputs Xenium Output Bundles (Figure 1) for each sample.
...
| Numbered figure captions |
|---|
| SubtitleText | Obtain the filtered H5 and Spatial imaging data outputs from the Space Ranger output Xenium Output Bundle on your machine |
|---|
| AnchorName | Obtain the filtered H5 and Spatial imaging data outputs from the Space Ranger output |
|---|
|  Image Removed
Image Removed| Xenium Output Bundle on your machine |
|
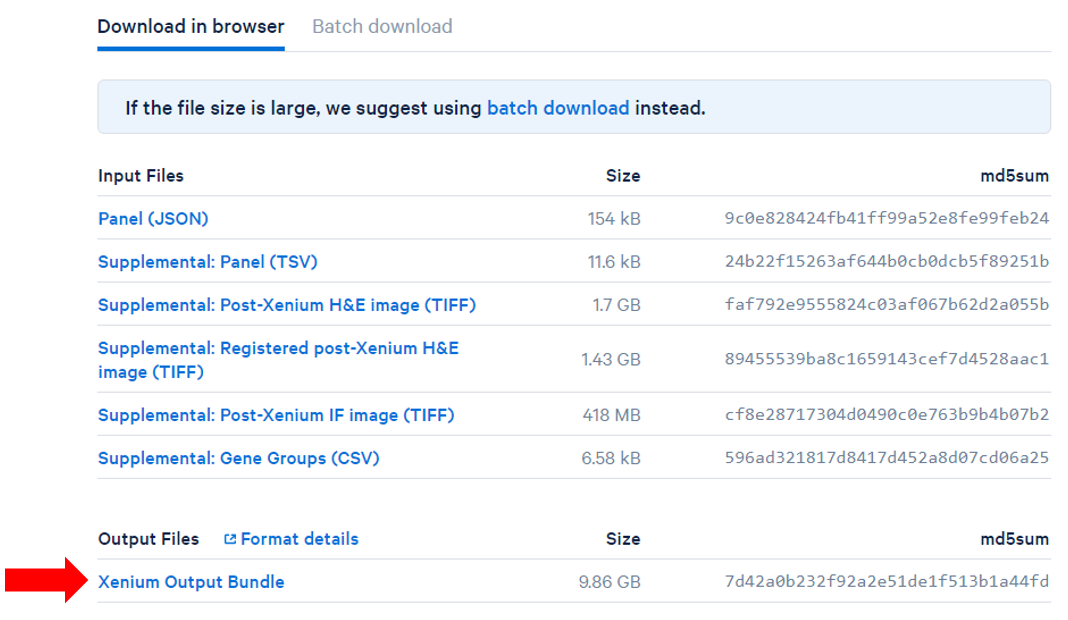 Image Added Image Added
|
- Navigate the options to select 10x Genomics Visium Space Ranger output Xenium Output Bundle as the file format for input.
Choose to import 10x Genomics Visium Space Ranger output Xenium for your project (Figure 2).
| Numbered figure captions |
|---|
| SubtitleText | Transfer files using the 10x Genomics Visium Space Ranger outputs Xenium importer |
|---|
| AnchorName | Transfer files during import |
|---|
|  Image Removed
Image Removed| using the 10x Genomics Xenium importer |
|
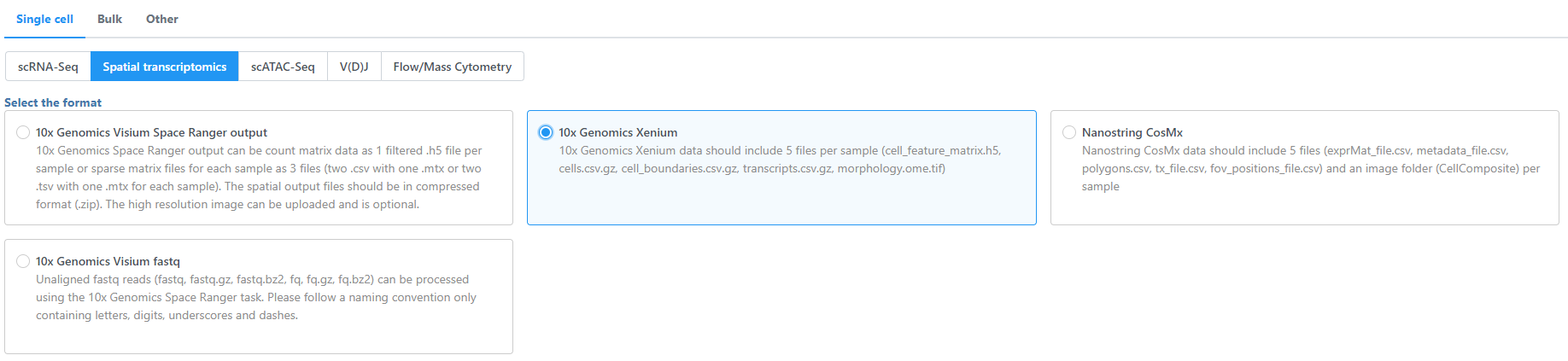 Image Added Image Added
|
- Click Transfer files on the homepage, under settings, or during import (Figure 3).
Proceed to transfer files as shown below using the 10x Genomics Visium Space Ranger outputs importer Xenium importer (Figure 3).
| Numbered figure captions |
|---|
| SubtitleText | Transfer files using the 10x Genomics Visium Space Ranger outputs Xenium importer |
|---|
| AnchorName | Transfer files during import |
|---|
|

|
- Navigate to the appropriate files for each sample (Figure 4). Please note that the 10x Genomics Space Ranger output can be count matrix data as 1 filtered .h5 file per sample or sparse matrix files for each sample as 3 files (two .csv with one .mtx or two .tsv with one .mtx for each sample). The spatial output files should be in compressed format (.zip). The high resolution image can be uploaded and is optional.
...
The Xenium output bundle should be included for each sample using Browse (Figure 4). The whole folder can be used, or a folder containing 5 files (cell_feature_matrix.h5, cells.csv.gz, cell_boundaries.csv.gz, transcripts.csv.gz, morphology.ome.tif) can be added per sample. Once added, the Cells and Features values will update.
| Numbered figure captions |
|---|
| SubtitleText | Add Visium Space Ranger outputsXenium output bundle |
|---|
| AnchorName | Navigate visium ouputsXenium import |
|---|
|
 Image Removed Image Removed Image Added Image Added
|
Once the download completes, the sample table will appear in the Metadata tab, with one row per sample (Figure 5).
...






