...
| Numbered figure captions |
|---|
| SubtitleText | Color the cells in the UMAP plot by their graph-based cluster assignment |
|---|
| AnchorName | UMAP of CITE-Seq data |
|---|
|
 Image Removed Image Removed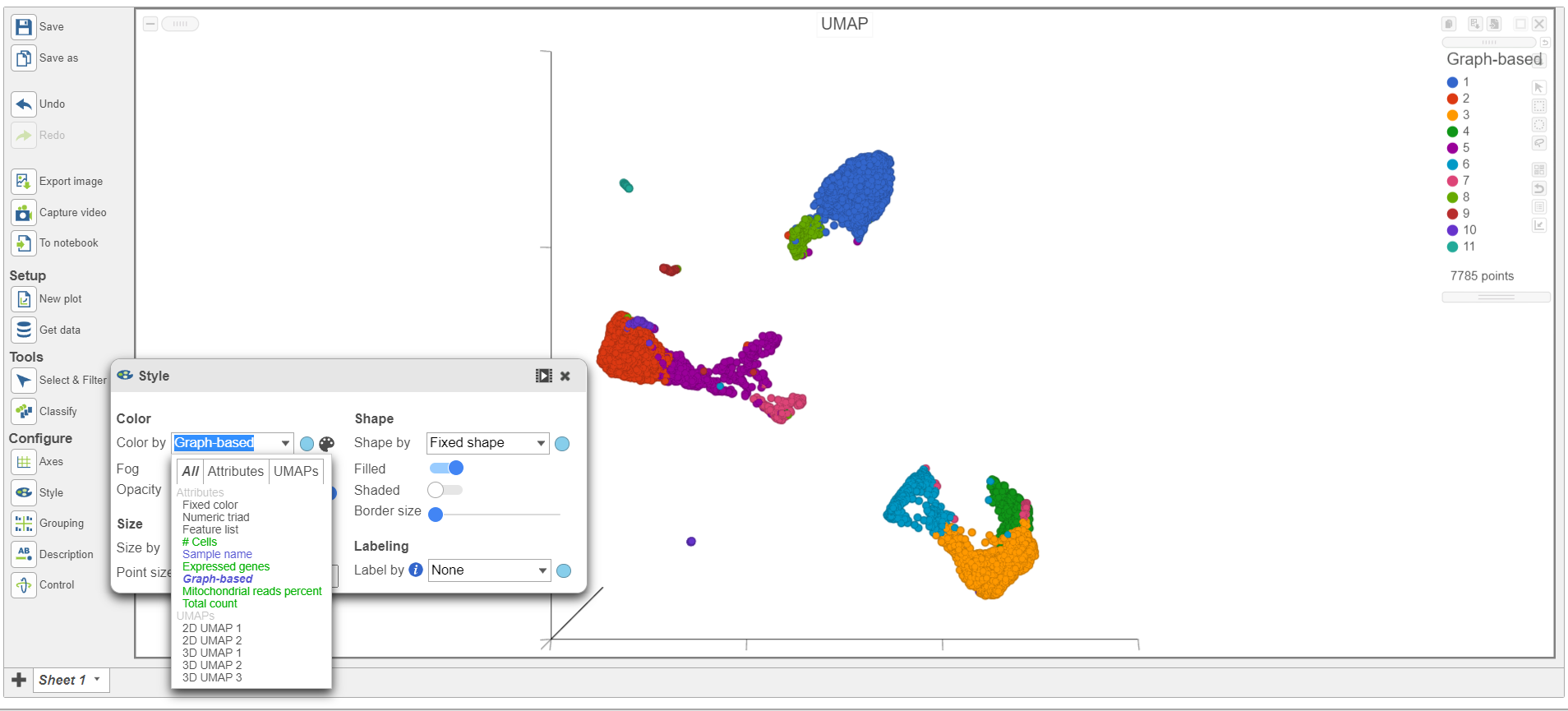 Image Added Image Added
|
The 3D UMAP plot opens in a new data viewer session (Figure 2). Each point is a different cell and they are clustered based on how similar their expression profiles are across proteins and genes. Because a graph-based clustering task was performed upstream, a biomarker table is also displayed under the plot. This table lists the proteins and genes that are most highly expressed in each graph-based cluster. The graph-based clustering found 11 clusters, so there are 11 columns in the biomarker table.
- Click and drag the 2D scatter plot icon from the Available plots card onto the New plot onto the canvas (Figure 2)
- Drop the 2D scatter plot to the right of the UMAP plot
...
| Numbered figure captions |
|---|
| SubtitleText | Add a 2D scatter plot and place it to the right of the UMAP plot |
|---|
| AnchorName | Add 2D scatter plot |
|---|
|
 Image Removed Image Removed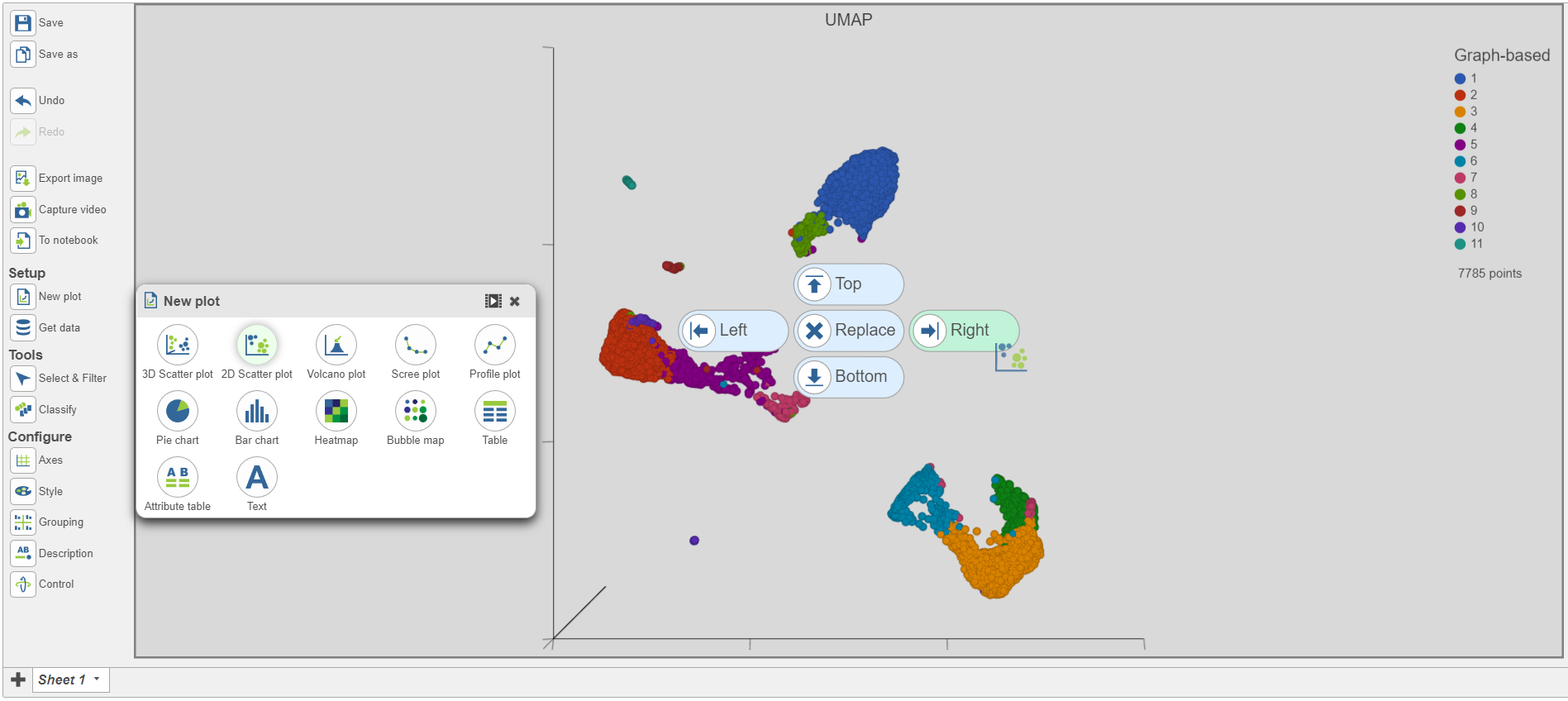 Image Added Image Added
|
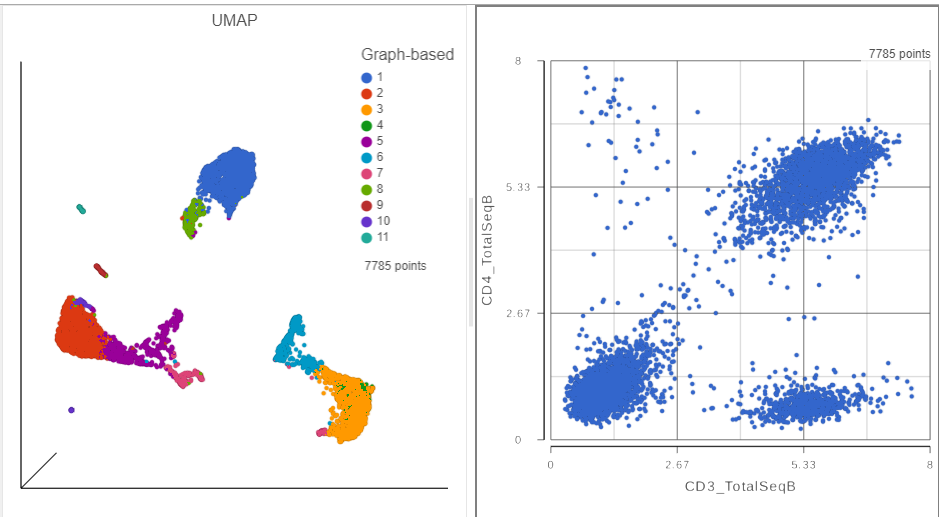
- Click Merged counts to use as data for the 2D scatter plot (Figure 3)
...
| Numbered figure captions |
|---|
| SubtitleText | The canvas now has a 2D scatter plot next to the UMAP |
|---|
| AnchorName | UMAP and 2D scatter plot |
|---|
|
|
- In the Selection card on the right, click Rule to In Select & Filter, click Criteria to change the selection mode
- Click the blue circle next to the Add rule drop-down menu (Figure 5)
...
| Numbered figure captions |
|---|
| SubtitleText | Click the blue circle to change the data source for the rule selector |
|---|
| AnchorName | Selection card rule mode |
|---|
|
 Image Removed Image Removed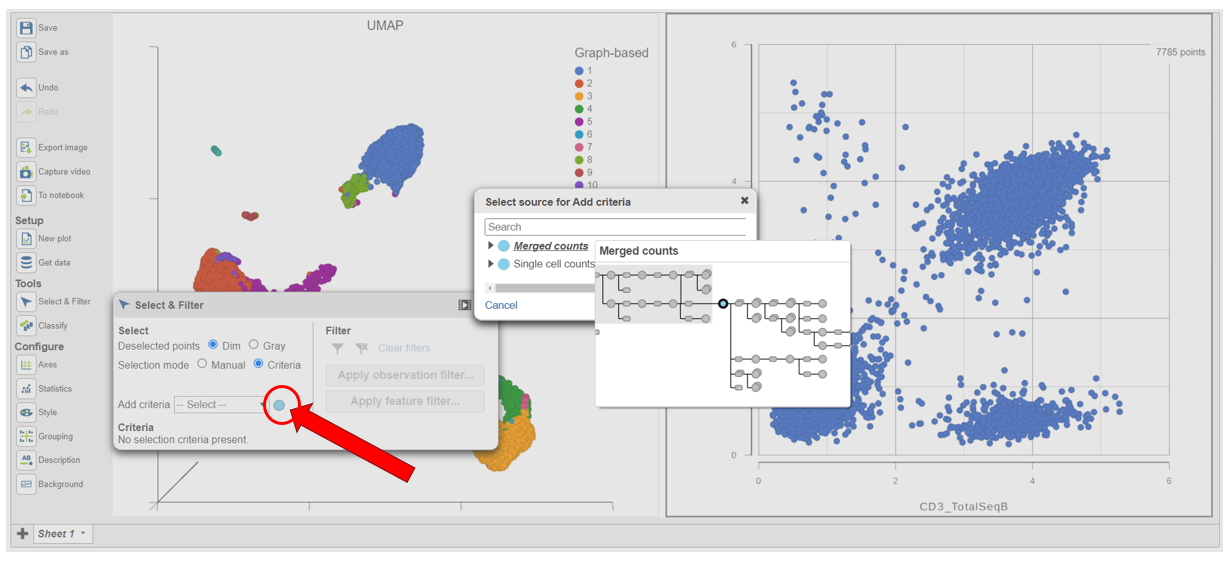 Image Added Image Added
|
- Click Merged counts to change the data source
- Choose CD3_TotalSeqB from the drop-down list (Figure 6)
...
| Numbered figure captions |
|---|
| SubtitleText | Choose the CD3_TotalSeqB protein marker as a selection rule |
|---|
| AnchorName | Choose CD3 Protein marker |
|---|
|
 Image Removed Image Removed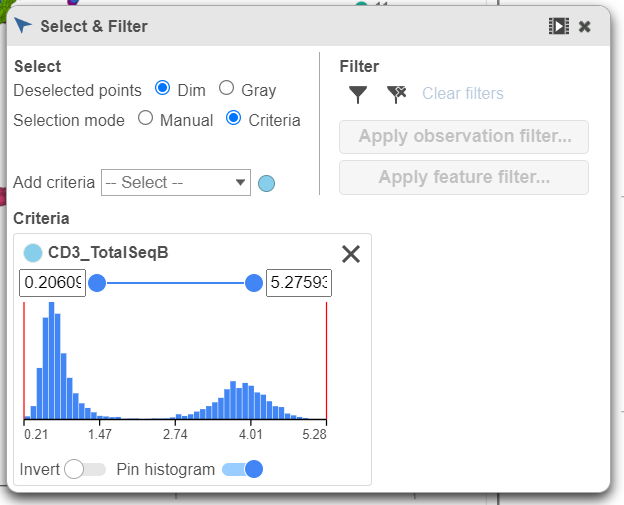 Image Added Image Added
|
- Click and drag the slider on the CD3D_TotalSeqB selection rule to include the CD3 positive cells (Figure 7)
...
| Numbered figure captions |
|---|
| SubtitleText | Use the slider to select cells with positive expression for the CD3 protein marker |
|---|
| AnchorName | Select CD3+ cells |
|---|
|
 Image Removed Image Removed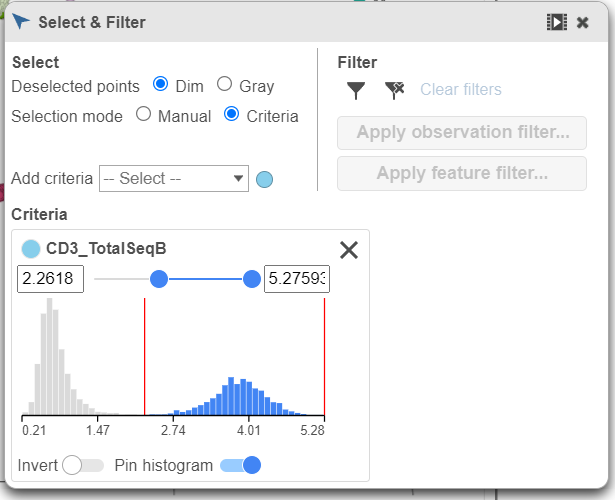 Image Added Image Added
|
As you move the slider up and down, the corresponding points on both plots will dynamically update. The cells with a high expression for the CD3 protein marker (a marker for T cells) are highlighted and the deselected points are dimmed (Figure 8).
...
| Numbered figure captions |
|---|
| SubtitleText | CD3+ cells are selected on both plots |
|---|
| AnchorName | CD3+ cells selected |
|---|
|
 Image Removed Image Removed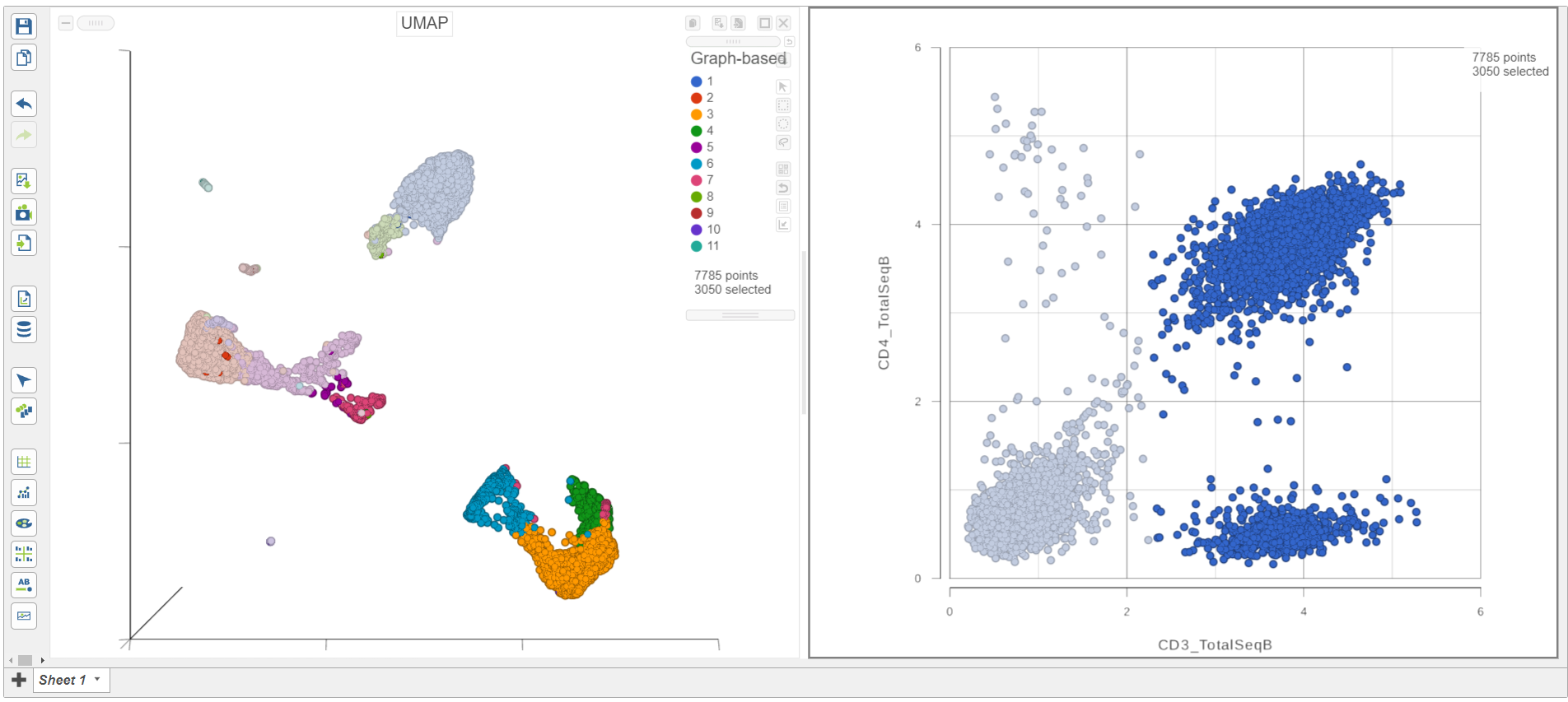 Image Added Image Added
|
- Click Merged counts in the Data card Get data on the left under Setup
- Click and drag CD8a_TotalSeqB onto the 2D scatter plot (Figure 9)
- Drop CD8_TotalSeqB onto the x-axis configuration option
...
| Numbered figure captions |
|---|
| SubtitleText | Change the feature plotted on the x-axis to CD8_TotalSeqB |
|---|
| AnchorName | CD8 protein on x-axis |
|---|
|
 Image Removed Image Removed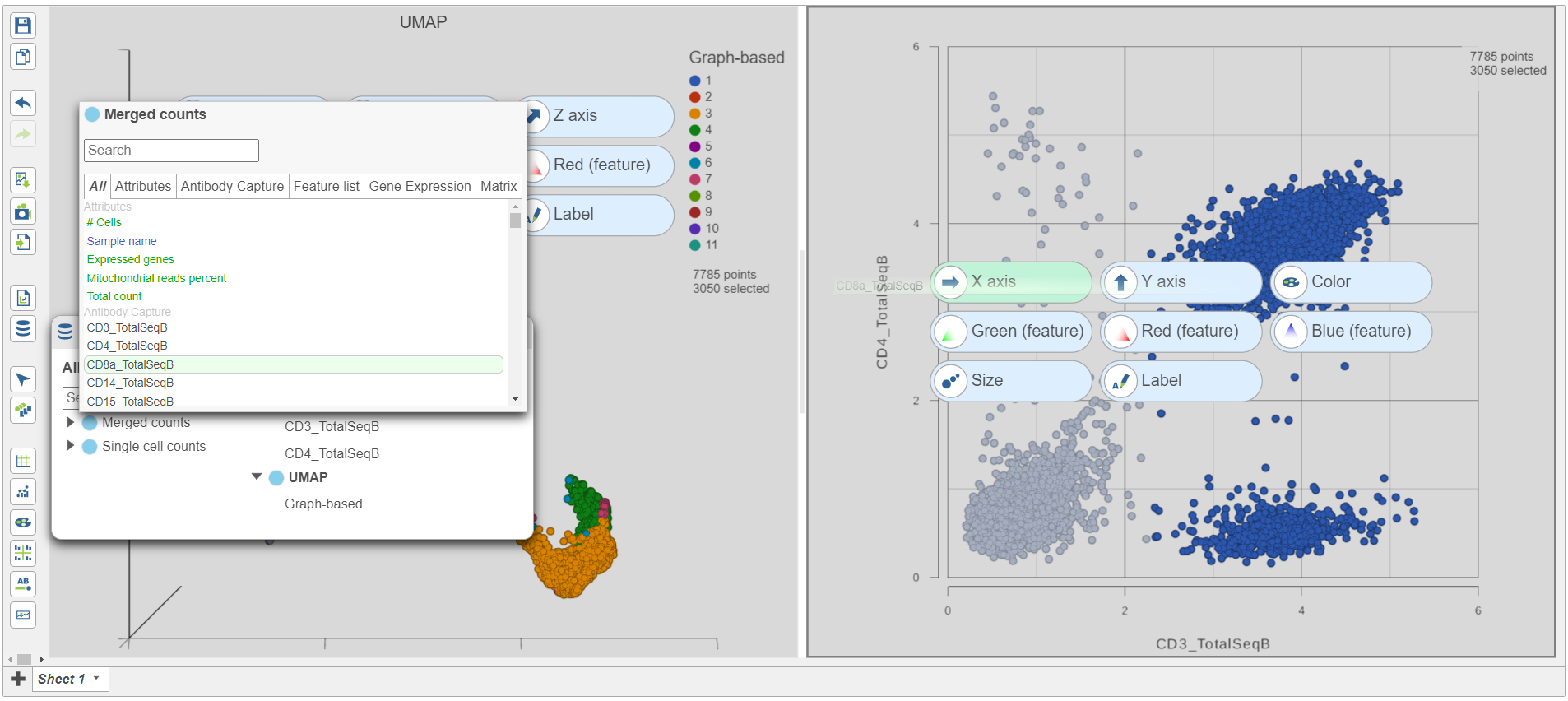 Image Added Image Added
|
The CD3 positive cells are still selected, but now you can see how they separate into CD4 and CD8 positive populations (Figure 10).
...
| Numbered figure captions |
|---|
| SubtitleText | Click the duplicate plot icon to make a copy of the 2D scatter plot |
|---|
| AnchorName | Duplicate plot |
|---|
|

|
- Click Merged counts in the Data card on the left Get Data icon under Setup
- Search for the CD4 gene
- Click and drag CD4 onto the duplicated 2D scatter plot
- Drop the CD4 gene onto the y-axis option
- Search for the CD8A gene
- Click and drag CD8A onto the duplicated 2D scatter plot
- Drop the CD8A gene onto the x-axis option
...
| Numbered figure captions |
|---|
| SubtitleText | Select the group of putative T cells |
|---|
| AnchorName | Lasso T cells |
|---|
|
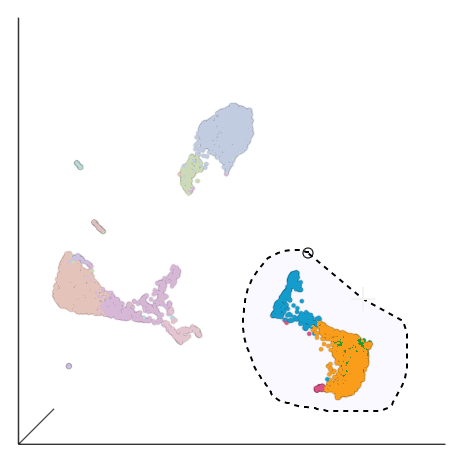
|
- Click
 in Filtering card on the right the Select & Filter tool to include the selected points
in Filtering card on the right the Select & Filter tool to include the selected points - Click
 in the top right of the plot to switch back to pointer mode
in the top right of the plot to switch back to pointer mode - Click and drag the plot to rotate it around
...
If you need to create more space on the canvas, hide the selection panel using the  Image Removedicon on the right and/or the
Image Removedicon on the right and/or the  Image Removed icon to hide the menu on the leftwords on the left using the arrow
Image Removed icon to hide the menu on the leftwords on the left using the arrow  Image Added.
Image Added.
| Numbered figure captions |
|---|
| SubtitleText | Resize plots to see more of the biomarker table |
|---|
| AnchorName | CITE-Seq biomarker table |
|---|
|
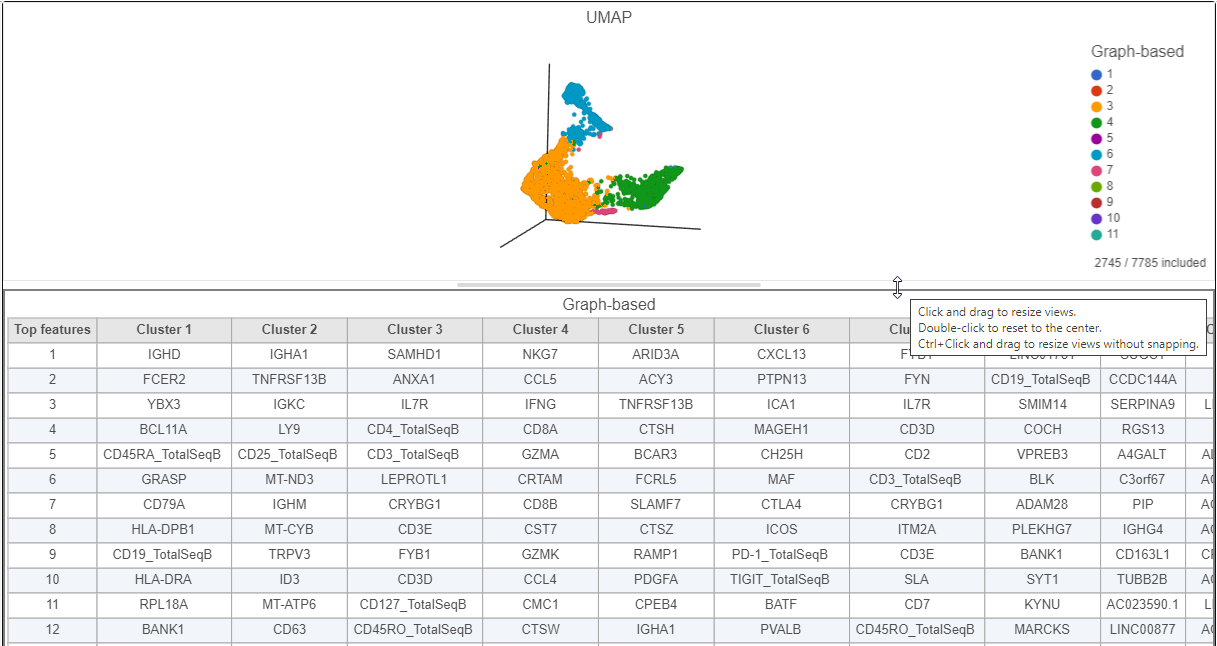
|
...
| Numbered figure captions |
|---|
| SubtitleText | The cells in the UMAP plot on the right are colored by their expression of CXCL13 (green) and NKG7 (red) marker genes. These cells belong to graph-based clusters 6 and 4, respectively, shown in the plot on the left |
|---|
| AnchorName | UMAP colored by CXCL13 and NKG7, respectively |
|---|
|
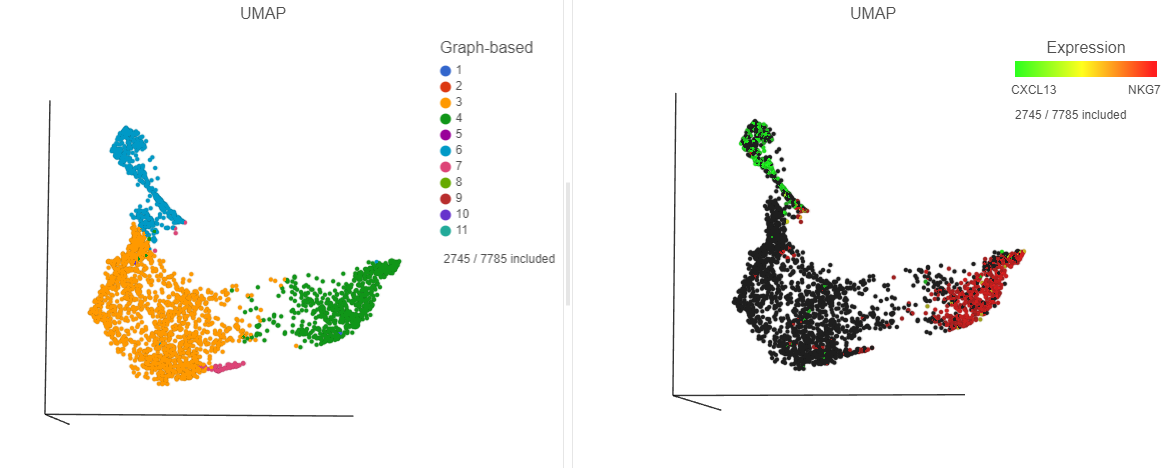
|
- In the Selection card on the right Select & Filter, click
 to remove the CD3_TotalSeqB filtering rule
to remove the CD3_TotalSeqB filtering rule - Click the blue circle next to the Add rule dropcriteria drop-down list
- Search for Graph to search for a data source
- Select Graph-based clustering (derived from the Merged counts > PCA data nodes)
- Click the Add rule dropcriteria drop-down list and choose Graph-based to add a selection rule (Figure 20)
...



















