Page History
...
Running Gene set enrichment
We recommend filtering to a set of genes you want to test for enrichment, but Gene set enrichment will run on any Feature list data node.Gene set enrichment task can be invoked on a differential analysis output (or filtered differential analysis output) data node or filtered count matrix data node. Since the data node including all the features will server as background, to get a meaningful result, always use a data node containing subset of features to invoke this task. Only gene names will be used in the computation.
- Click a Feature list data node
- Click the Biological interpretation section of the toolbox
- Click Gene set enrichment
- Configure the background gene list (optional)
The background gene list is used as the list of possible genes. By default, this is the genes included in the selected gene set database. If your assay limits the genes that could be detected, you may want to specify a background list.
- Choose the Gene set
...
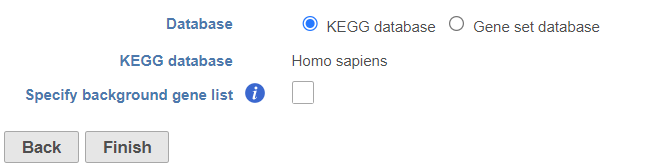
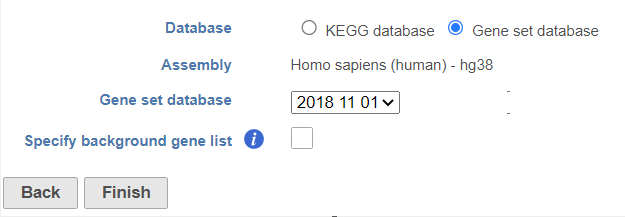
- There are two options for Database. KEGG database requires special license (Figure 1).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
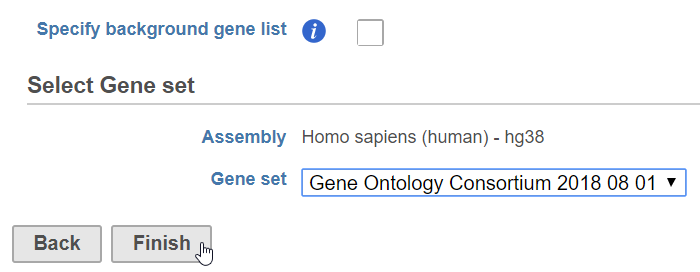
- When choose Gene set database, it is user defined database, see the details in the Adding a Gene Set chapter. The gene sets available for the current Assembly are listed under the Gene set database drop-down list (Figure 2). The assembly is automatically selected, if possible. If the assembly cannot be detected, you can specify it using a drop-down menu.
- Click Finish to run (Figure 1)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
By default, the groups are defined by Gene Partek distribute Gene Ontology (GO) for human and mouse genomes, a bioinformatics initiative to unify the representation of gene and gene product attributes across various species [1, 2].
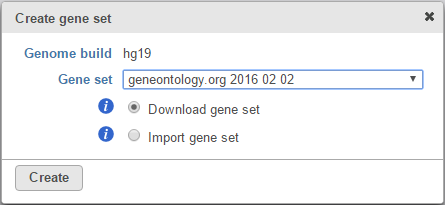
Alternatively, selecting the Add gene ontology source from the Gene set drop down list option opens another dialog (Figure 3), where you can either Download gene set from Partek® (Recent GO database gene sets for human, mouse and rat are available) or Import gene set. The latter option takes you to the file browser, where you can point to the file that you want to use (not shown). Partek® Flow® accepts .gmt files as gene set inputs.
| Numbered figure captions | ||||||||
|---|---|---|---|---|---|---|---|---|
| ||||||||
- Configure the background gene list (optional)
The background gene list is used as the list of possible genes. By default, this is the genes included in the selected gene set database. If your assay limits the genes that could be detected, you may want to specify a background list.
- Click Finish to run
The result is stored under an Enrichment task node. To open it, double click on the node or select the respective Task report from the context sensitive menu.
...
Task report
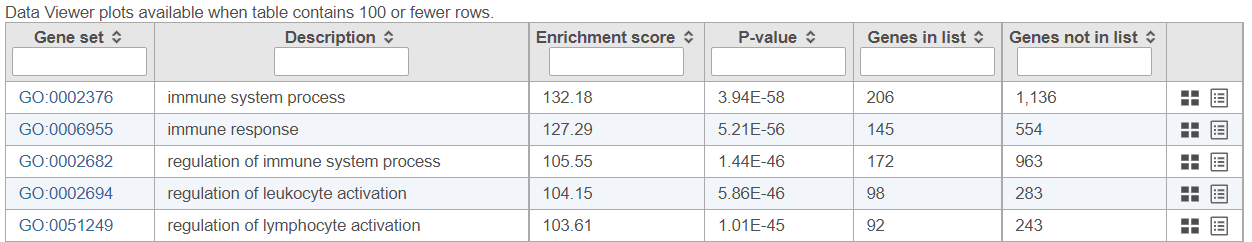
Figure 4 3 shows an example Gene set enrichment task report using GO database. The table contains one gene set per row (Gene set column; the column entries are hyperlinks when using the distributed GO gene sets), with the category name in the Description column. The categories are ranked by the Enrichment score, which is the negative natural logarithm of the enrichment p-value (P-value column) derived from Fisher's exact test on the underlying contingency table. The higher the enrichment score, the more overrepresented the GO category is within the input list of significant genes. The columns can be searched by typing in the search term in the respective box (and hitting Enter), or sorted by selecting the double arrow icon ( ).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
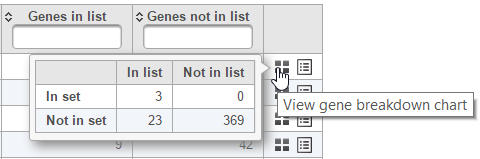
The contingency table (Figure 54) can be displayed by selecting the View gene breakdown chart icon on the right (). The term "list" refers to the list of significant genes, while the term "set" refers to the respective GO category. The first row of the contingency table is also seen in the report, namely the Genes in list and Genes not in list columns.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
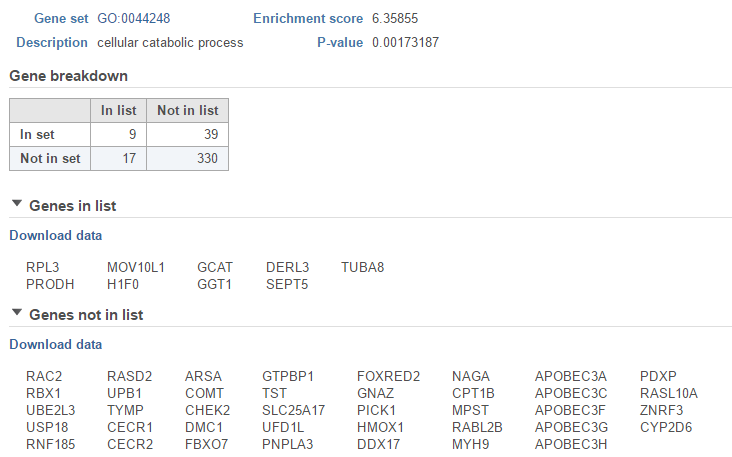
The View extra details () button provides additional information on the GO category (Figure 65). In addition to the details already given in the report, a full list of Genes in list and Genes not in list can be inspected and downloaded (Download data) to the local computer as a text file.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
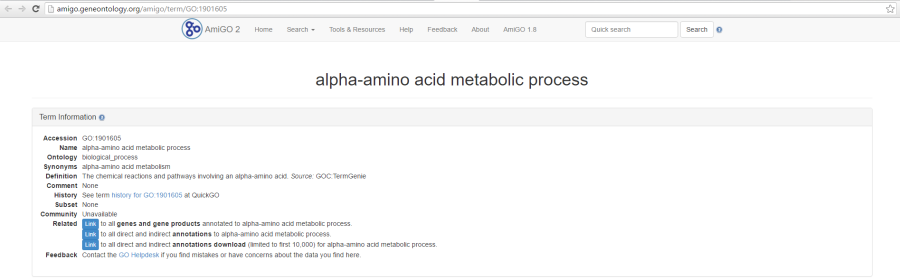
As previously mentioned, if you are using the GO gene sets distributed by Partek, the GO identifiers in the first column are hyperlinks to the Gene Ontology web-site entries (an example shown in Figure 76).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Interactive KEGG pathway maps
When KEGG database is used, on the enrichment task report, when click on a pathway ID in the Gene set column, a KEGG pathway gene network picture is displayed (Figure 7)
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Each rectangle on the map represents a gene product in the pathway. Gene products are mostly proteins coded by a gene or group of genes, but they could be RNA too. Related pathways are shown as large rounded rectangles. Chemical compounds, DNA or other molecules are shown as circles.
Coloring the map
The pathway map is colored by the first fold-change column in the input Feature list data node. The control panel on the left can be used to configure the colors of the pathway map. In all options, rectangles colored white do not have gene information. Options for coloring include:
- Fixed color: all genes are colored black.
- Genes in list: all genes in the list are colored, default color is yellow, but this can be configured. Genes not in the list are black.
- Statistics in the gene list: .e.g FDR, p-value, Fold change etc. Colors can be customized by clicking on the color square to change.
Interactive features
Mousing over a rectangle shows the genes indicated by the rectangle in the tooltip (Figure 8). Genes are listed on rows with all aliases in the KEGG database included on the row. Genes that are in the list are shown in bold. The gene being used to color the rectangle is shown in red.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
On KEGG pathway maps that include chemical compounds, the chemical structure is shown in the tooltip on mouse-over (Figure 10).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
Clicking a rectangle opens the page for that gene or group of genes on the KEGG website in a new tab in your web browser.
Click the Save image icon to download a PNG file showing the configured KEGG pathway map to your local computer.
Visualizing gene set enrichment results
If the gene set enrichment table has fewer than 100 results (rows), the GO categories can be visualized in the Data Viewer. Otherwise, a notification is displayed in the top left corner (Figure 711).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
...
If needed, filter down the number results, for instance by using a cut-off based on the enrichment score. Type in the cut-off value in the text box beneath the Enrichment score and hit enter (an example is shown in Figure 812). Once the number or results falls below 100, a link to the Data Viewer will be displayed (Figure 8). Click on the View plots in Data Viewer link to open a new Data Viewer session.
...
Two plots are loaded into Data Viewer (Figure 913). Both plots show enrichment score on the horizontal axis and gene ontology categories (i.e. the ones present in the gene enrichment table) on the vertical axis. The plots show enrichments scores (Enrichment score column of the gene ontology table) and - in addition - the plot on the left uses color range to depict enrichment P-value (green = low, red = high P-value).
...