Page History
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
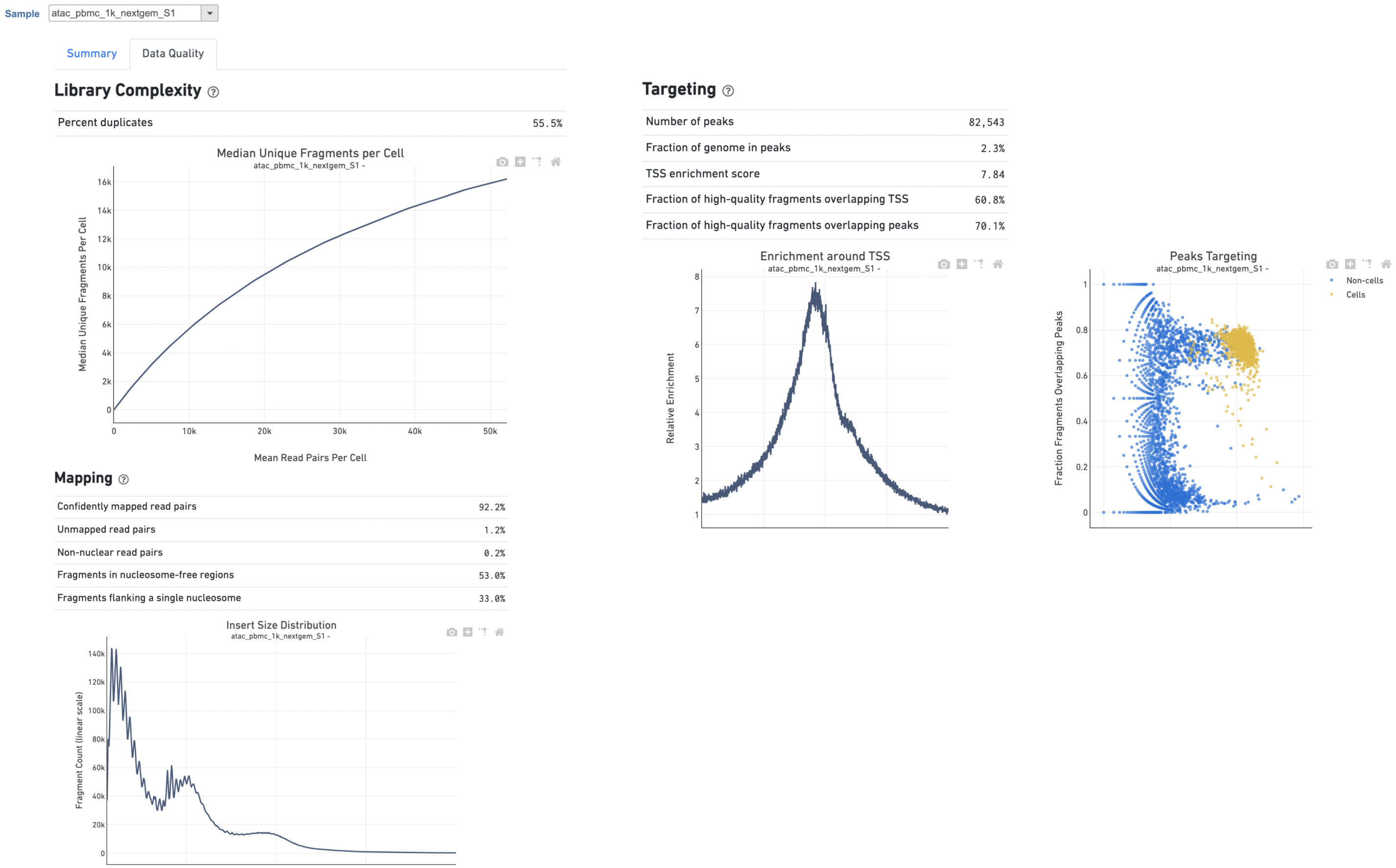
Other parameter to adjust (Figure 2):
Subsample percentile: Downsample to preserve this fraction of reads
Users can click Configure to change the default settings In in Advanced options (Figure 42).
Include introns: Count reads mapping to intronic regions. This may improve sensitivity for samples with a significant amount of pre-mRNA molecules, such as nuclei.
Expected cells: Expected number of recovered cells. Default: 3,000 cells.
Force cells: Force pipeline to use this number of cells, bypassing the cell detection algorithm. Use this Override peak caller: To override the peak caller, users specify peaks to use in downstream analyses from supplied 3-column BED file. The supplied peaks file must be sorted by position and not contain overlapping peaks; comment lines beginning with `#` are allowed.
Force cells: Define the top N barcodes with the most fragments overlapping peaks as cells and override the cell calling algorithm. N must be a positive integer <= 20,000. Use this option if the number of cells estimated by Cell Ranger -ATAC is not consistent with the barcode rank plot.
Memory limit (GB): Restricts Cell Ranger - Gene ExpressionATAC to use specified amount of memory (in GB) to execute pipeline stages.
...