...
| Numbered figure captions |
|---|
| SubtitleText | Space Ranger task in the toolbox |
|---|
| AnchorName | space_ranger_menu |
|---|
|
 Image Removed Image Removed
|
...
 Image Added Image Added
|
For both Space Ranger or Cell Ranger, you will need to create a Cell Ranger reference tasks, a Reference assembly is required (Figure 2). To define the Reference assembly, first select the Genome build for the organism of interest, then select the annotation Index. In this manner, custom libraries can be created (e.g. keep the Genome build but change the annotation Index).
| Numbered figure captions |
|---|
| SubtitleText | Space Ranger setup dialog. If no Cell Ranger reference is present, one needs to be created first |
|---|
| AnchorName | space_ranger_setup |
|---|
|
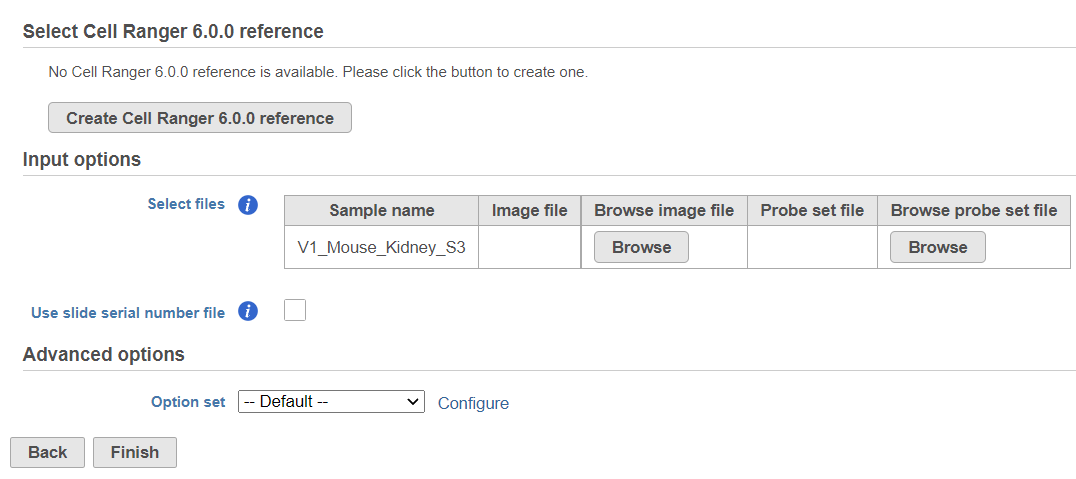 Image Removed Image Removed Image Added Image Added
|
To create add a new reference, push the Create Cell Ranger 6.0.0 reference button and pick the appropriate Cell Ranger reference name new species genome, choose New assembly from the drop down for Genome build which will open a new window with configuration options to edit, then click Create (Figure 3) and then push Create.
| Numbered figure captions |
|---|
| SubtitleText | Downloading a Cell Ranger reference. Supported references appear in the drop down menu |
|---|
| AnchorName | download_cell_ranger_reference |
|---|
|
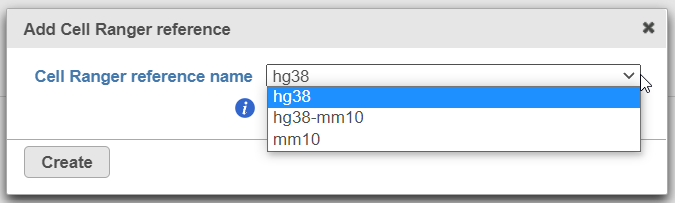 Image Removed Image Removed
|
Downloading a reference is a separate task and may take some time. However, the reference will become immediately available in the Cell Ranger 6.0.0. reference dropdown (Figure 4).
| Numbered figure captions |
|---|
| SubtitleText | Selecing Cell Ranger reference from the drop down list |
|---|
| AnchorName | selec_cell_ranger_reference |
|---|
|
 Image Removed Image Removed
|
Alternatively, Cell Ranger reference can be added beforehand, to the Library file management page (on the Other library files tab). At the moment, the following references are available for download from Partek: hg38, mm10, and hg38-mm10 (chimaeric genome).
 Image Added Image Added
|
The sample table under Input options has one row per sample (Figure 2). Image file is required, and that is a single hematoxylin and eosin brightfield image in either .jpg or .tiff format. Click on the Browse button under Browse image file and the file browser will come up. Point to the image file and push Continue. The Formalin-fixed paraffin-embedded (FFPE) image files require the Probe set file otherwise it is optional; it is a .csv file specifying the probe set used (=target panel).
If you want to specify the sample's slide and area information, tick mark the box by the Use slide serial number file under Advanced options and then click Browse to point to the file. The file should be tab-delimited with samples on rows. The first column is the sample name, the slide name is on the second column, slide area is on the third column.
...
| Numbered figure captions |
|---|
| SubtitleText | Advanced options of Space Ranger |
|---|
| AnchorName | advanced_space_ranger_options |
|---|
|
 Image Modified Image Modified
|
Space Ranger task report
...
| Numbered figure captions |
|---|
| SubtitleText | Space Ranger report. The report matches the Summary HTML from the Space Ranger pipeline by 10x Genomics. Each sample is shown as a separate page, with two tabs - Summary and Analysis. If a project contains more than one sample, sample selector will be displayed in top left |
|---|
| AnchorName | space_ranger_HTML_report |
|---|
|
 Image Modified Image Modified
|
Annotate Visium image
...
| Numbered figure captions |
|---|
| SubtitleText | Annotate Visium image setup page |
|---|
| AnchorName | annotate_visium_image_setup |
|---|
|
 Image Modified Image Modified
|
You can find the location of the <project-name>_spatial.zip file using the following steps. Select the Space Ranger task node (i.e. the rectangle) and then click on the Task Details (toolbox). Click on the Output files link to open the page with the list of files created by the Space Ranger task. Mouse over any of the files to see the directory in which the file is located. Figure 9 shows the path to the .zip file which is required for Annotate Visium image.
...
| Numbered figure captions |
|---|
| SubtitleText | Mousing over a file on the Output files page shows a baloon with the file location |
|---|
| AnchorName | file_location |
|---|
|
 Image Modified Image Modified
|
Annotate Visium image task creates a new node, Annotated counts. Double click on the Annotated counts node to invoke the Data Viewer showing data points overlaid on top of the microscopy image (Figure 10).
...
| Numbered figure captions |
|---|
| SubtitleText | Data Viewer session as a result of opening an Annotated counts data node. Each data point is a tissue spot |
|---|
| AnchorName | data_viewer_annotated_counts |
|---|
|
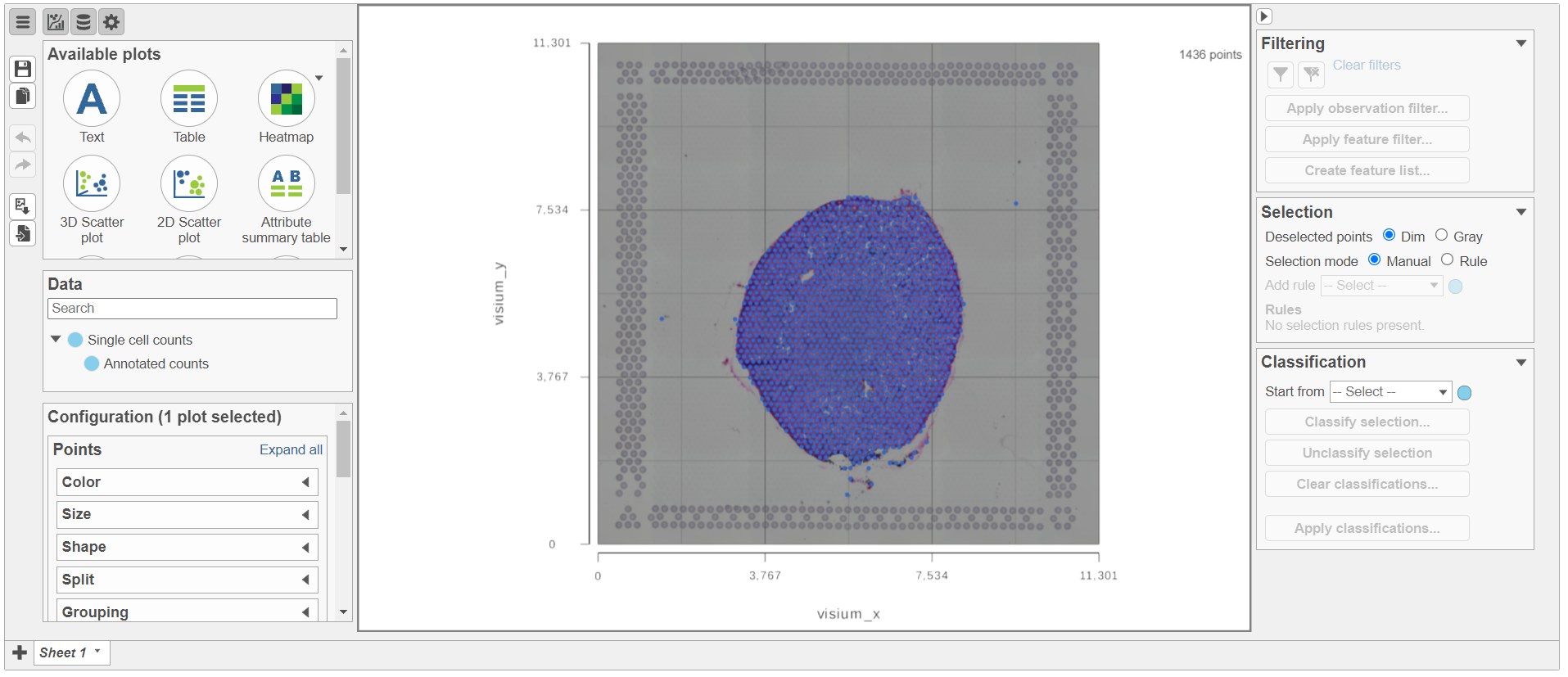 Image Modified Image Modified
|
Resources
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
...










