Page History
...
To run the Cell Ranger - Gene Expression task for scRNA-seq data in Flow, select Unaligned reads datanode, then select Cell Ranger - Gene Expression in the 10x Genomics section (left panel, Figure 1). For Feature Barcode data, there will be two datanodes data nodes once the fastq files have been imported in into Flow properly - mRNA and protein (right panel, Figure 1). Users should select mRNA datanode to trigger the Cell ranger - Gene Expression task.
...
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
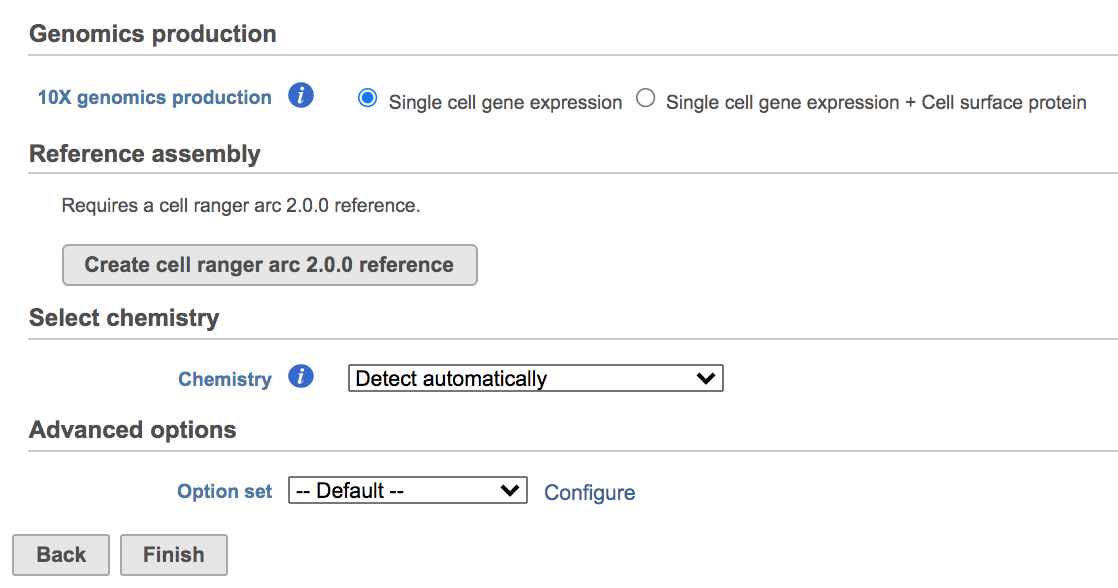
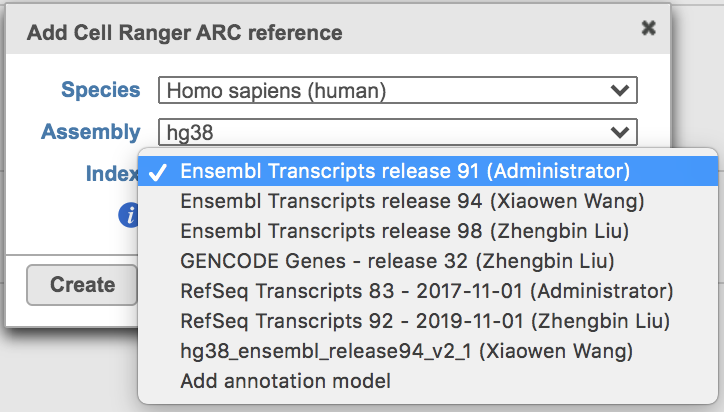
Clicking the big grey button of Create Cell Ranger ARC 2.0.0 reference would pop up a new window where lists the requirements that users need to fill in (Figure 3). To create the same reference genomes (2020-A) that are provided in Cell Ranger by default, the transcriptome annotations are respectively GENCODE v32 for human and vM23 for mouse, which are equivalent to Ensembl 98[3]. If users don't have options in the dropdown list, they can click Add annotation model (GTF file) for Index, or New assembly... (FASTA file)for Assembly and upload the files.
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
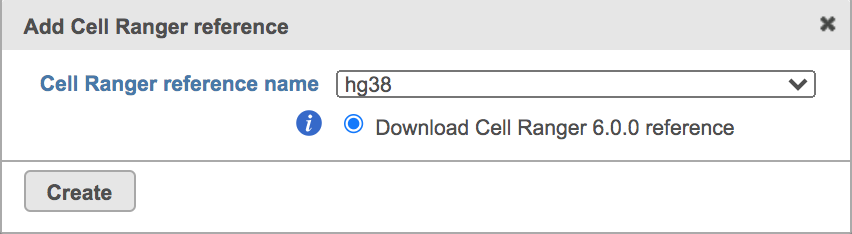
Once the right options has been chosen/provided, simply press the Create button to finish. The reference assembly of ‘Homo sapiens (human) - hg38’ has been created as an example here (Figure 4).
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
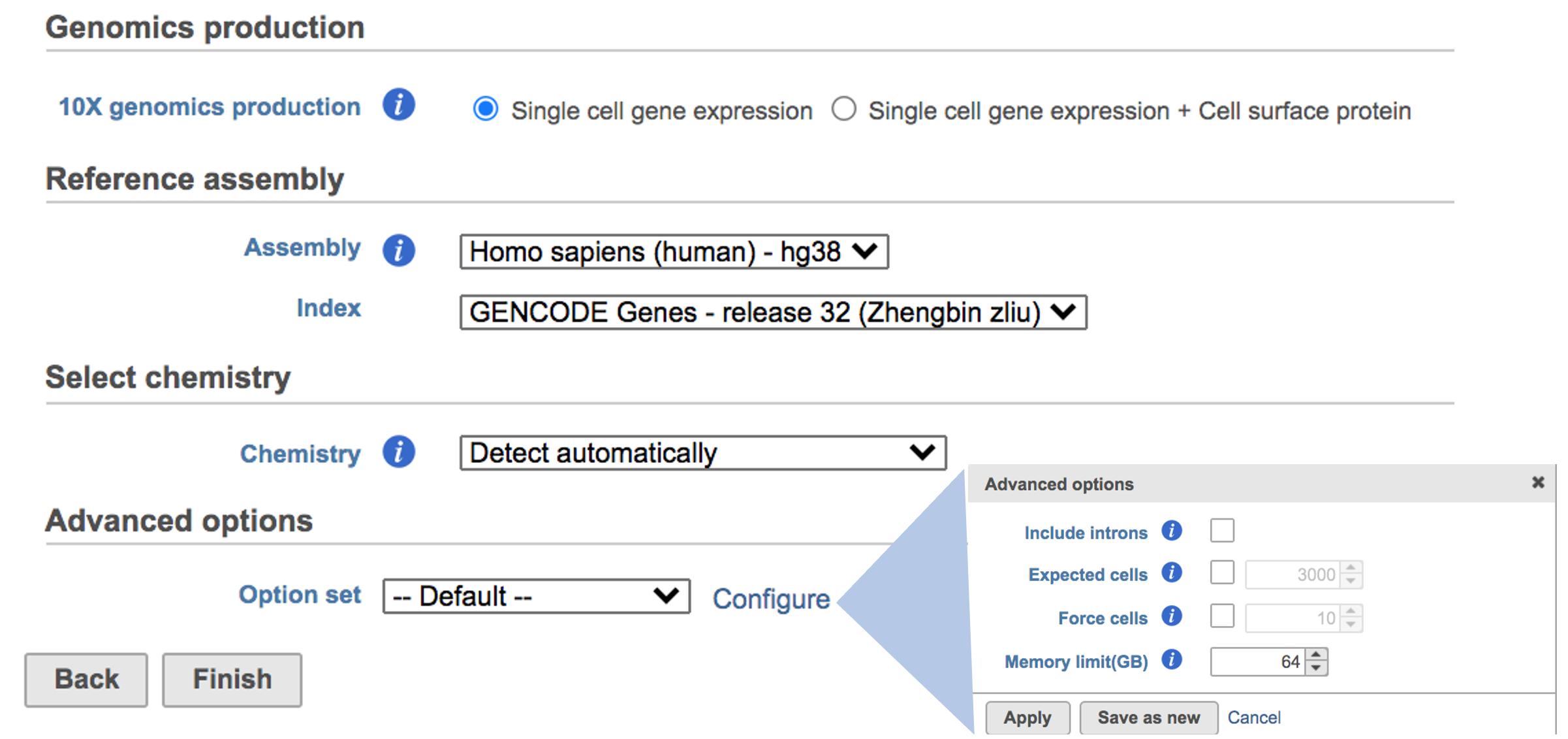
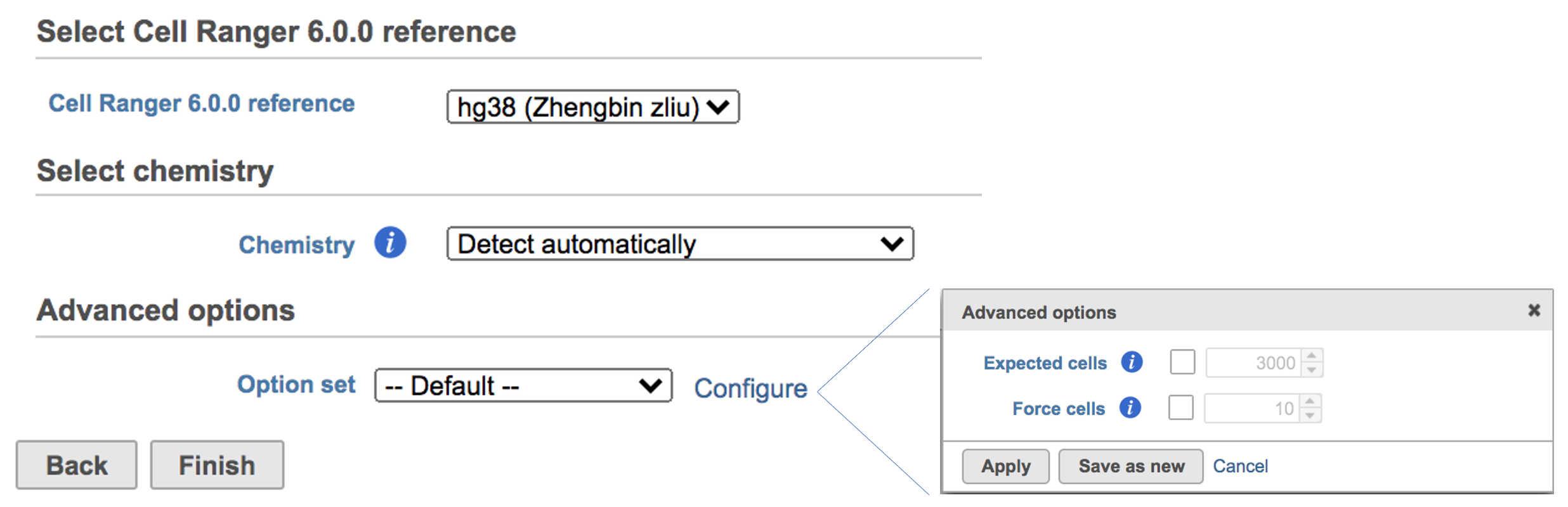
The main task menu will be refreshed as below above (Figure 5) for gene expression data if references have been added. After the correct reference has been selected, users Users can go ahead click the Finish button to run the task as default.
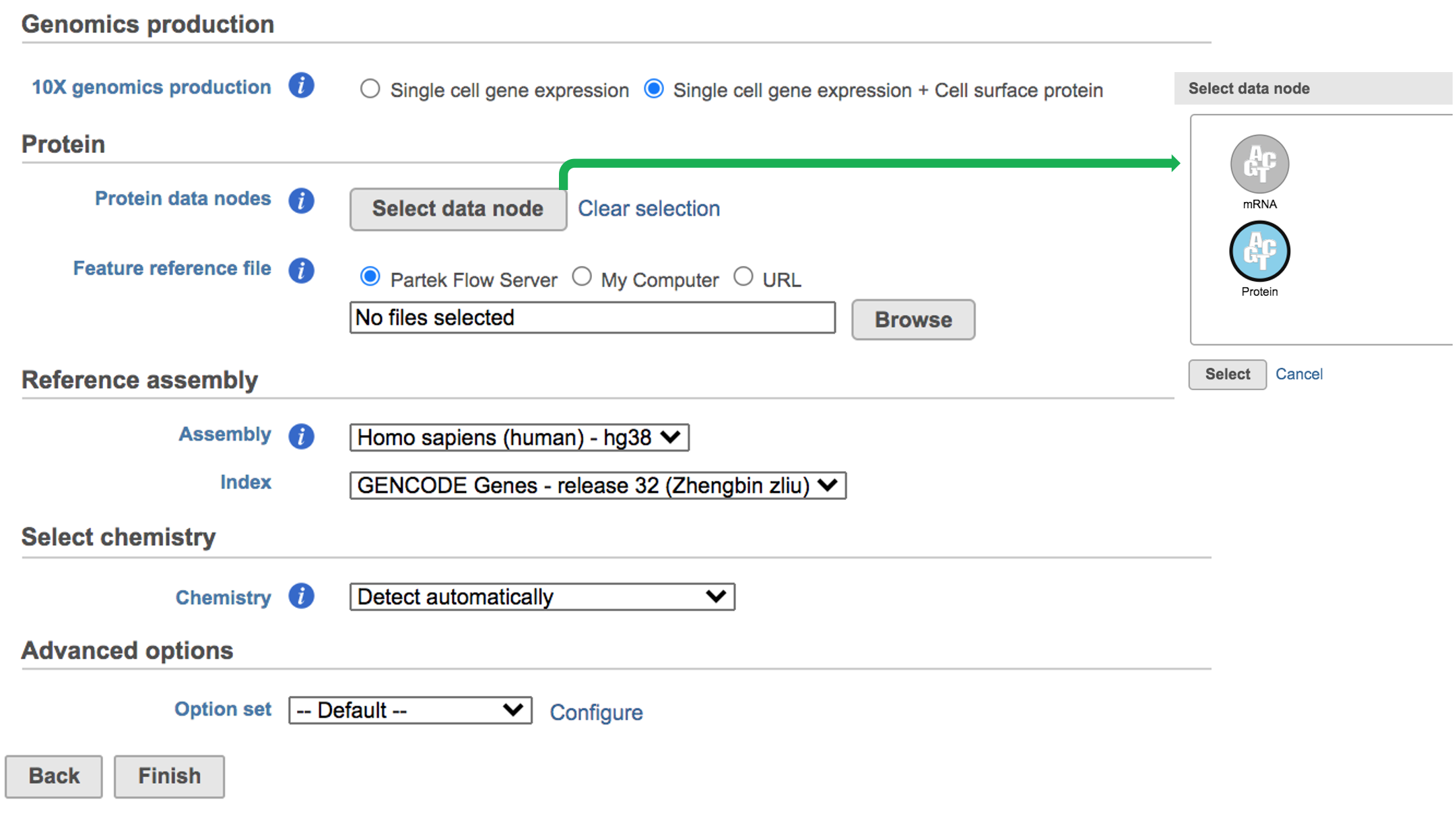
While for Feature Barcode data, there's are more information needed besides reference assembly. An additional section of Protein has been added to the interface if Single cell gene expression + Cell surface protein has been selected for Feature Barcode data (Figure 5). Firstly, push the button Select data node and select the correct data for feature of antibody capture or protein in the newly popped-out window (top right, Figure 5). Then upload the feature reference file (.csv) prepared by users for their datasets. A Feature Reference CSV file declares the molecule structure and unique Feature Barcode sequence of each feature present in your experiment. It should include at least six columns: id, name, read, pattern, sequence and feature_type. An example of TotalSeq™-B Feature Reference CSV has been linked here. But for more details, please refer to 10x Genomics webpage[4].
| Numbered figure captions | ||||
|---|---|---|---|---|
| ||||
A new data node named ‘Single cell counts’ will be displayed in Flow if the task is running (Figure 6). This data node contains filtered feature barcode count matrix. To open the task report when the task is finished, double click the output data node, or select the ‘Task report’ in the section after single clicking the data node. Users then will find the task report (Figure 7) is the same to the ‘Summary HTML’ from Cell Ranger output.
...
https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome
https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/6.0/release-notes
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/4.0/release-notes
- https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/feature-bc-analysis#feature-ref
| Additional assistance |
|---|
| Rate Macro | ||
|---|---|---|
|
...